Abstract
A number of clinical trials have shown that KRAS mutations of colorectal cancer (CRC) can predict a lack of responses to anti-epidermal growth factor receptor–based therapy. Recently, there have been several studies to elucidate metabolism reprogramming in cancer. However, it remains to be investigated how mutated KRAS can coordinate the metabolic shift to sustain CRC tumor growth. In this study, we found that KRAS mutation in CRC caused alteration in amino acid metabolism. KRAS mutation causes a marked decrease in aspartate level and an increase in asparagine level in CRC. Using several human CRC cell lines and clinical specimens of primary CRC, we demonstrated that the expression of asparagine synthetase (ASNS), an enzyme that synthesizes asparagine from aspartate, was upregulated by mutated KRAS and that ASNS expression was induced by KRAS-activated signaling pathway, in particular PI3K-AKT-mTOR pathway. Importantly, we demonstrated that KRAS-mutant CRC cells could become adaptive to glutamine depletion through asparagine biosynthesis by ASNS and that asparagine addition could rescue the inhibited growth and viability of cells grown under the glutamine-free condition in vitro. Notably, a pronounced growth suppression of KRAS-mutant CRC was observed upon ASNS knockdown in vivo. Furthermore, combination of L-asparaginase plus rapamycin markedly suppressed the growth of KRAS-mutant CRC xenografts in vivo, whereas either L-asparaginase or rapamycin alone was not effective. These results indicate ASNS might be a novel therapeutic target against CRCs with mutated KRAS.
Abbreviations: CRC, colorectal cancer; ASNS, asparagine synthetase
Introduction
Mutations in the KRAS gene are found in various types of cancer including pancreatic ductal cell adenocarcinoma (PDCA) and colorectal cancer (CRC). A number of clinical trials have shown that KRAS mutations in CRC can predict a lack of responses to the anti-epidermal growth factor receptor (EGFR)–based therapy. The use of anti-EGFR antibodies, cetuximab and panitumumab, is now limited to patients with KRAS wild-type CRC [1], [2], [3]. Therefore, the development of new therapy for CRCs with mutated KRAS has been desired clinically.
In recent years, there has been intense interest to understand the reprogramming of metabolism in cancer [4], [5], [6], [7]. One of the metabolic hallmarks of malignant tumor cells is their dependency on aerobic glycolysis, known as the Warburg effect [4], [5]. The role of KRAS signaling in the regulation of aerobic glycolysis has been reported in several types of cancer, although the molecular mechanism behind the upregulation of glucose metabolism is yet to be elucidated. For example, in a PDCA mouse model, mutated KRAS was shown to maintain tumor growth by stimulating glucose uptake and channeling glucose intermediates into the hexosamine biosynthesis pathway (HBP) and pentose phosphate pathway (PPP) [8]. Notably, knockdown of rate-limiting enzymes in HBP or PPP suppressed tumor growth, indicating their potential as therapeutic targets. In CRC cells, the increase of glucose transporter 1 (GLUT1) expression and glucose uptake was critically dependent on KRAS or BRAF mutations [9]. Fluorodeoxyglucose (FDG) positron emission tomography scans are used to evaluate glucose metabolism by measuring the uptake of FDG, a glucose analog. We previously reported that CRC cells with mutated KRAS increased FDG accumulation by upregulation of GLUT1 [10], [11], [12]. However, it remains to be investigated how mutated KRAS can coordinate the metabolic shift to sustain tumor growth and whether specific metabolic pathways are essential for the KRAS mutation-mediated tumor maintenance in CRC.
In addition to their glucose dependency, malignant cells rely on glutamine to support cell growth and survival [13], [14]. Glutamine is one of the most heavily consumed nutrients by cells in culture and the most abundant amino acid in circulation [15]. Once imported into the cells, glutamine serves as a carbon source for the tricarboxylic acid (TCA) cycle and a nitrogen source for nucleotide and nonessential amino acids. In purine and pyrimidine biosynthesis, glutamine donates its amino group and is subsequently converted to glutamate. In turn, glutamate serves as the primary nitrogen source for other nonessential amino acids by providing the amino group and is subsequently converted to α-ketoglutarate. The glutamine-derived α-ketoglutarate replenishes the TCA cycle by providing oxaloacetate that condenses with acetyl-CoA to maintain the TCA cycle and support de novo fatty acid biosynthesis. In addition to providing carbons and nitrogens for biosynthesis, glutamine is also involved in other cellular processes, including antioxidative stress and the mammalian target of rapamycin (mTOR) signaling. The spectrum of glutamine-dependent tumors and the mechanisms by which glutamine supports cancer metabolism are being actively investigated [13], [14], [15], [16], [17], [18]. In the PDCA mouse model, glutamine supports the growth of pancreatic cancer through an oncogenic KRAS-regulated metabolic pathway [19]. Most cells utilize glutamine dehydrogenase (GLUD1) to convert glutamine-derived glutamate into α-ketoglutarate in the mitochondria to fuel the TCA cycle. However, PDCA cells rely on a distinct pathway in which glutamine-derived aspartate is transported into the cytoplasm and converted to oxaloacetate by aspartate transaminase (GOT1). In glioma and neuroblastoma, asparagine plays a critical role in regulating cellular adaptation to glutamine depletion [20]. Asparagine was shown to be necessary and sufficient to suppress apoptosis induced by the glutamine withdrawal without restoring the levels of other nonessential amino acids or TCA cycle intermediates. Asparagine synthetase (ASNS), the enzyme that synthesizes de novo asparagine from aspartate and glutamine, was required to suppress glutamine withdrawal–induced apoptosis, and its expression was statistically correlated with poor prognosis.
The present study aimed to investigate how mutated KRAS could regulate metabolic reprograming in CRC and whether metabolic enzymes associated with mutated KRAS could be novel therapeutic targets for CRC with KRAS mutations. Given that cancer cells rely on changes in metabolism to support their growth and survival, targeting the metabolism is a potential cancer treatment strategy. There are a few reports regarding KRAS mutation-related metabolic alterations in CRC. Here, we revealed that mutated KRAS upregulated ASNS expression through the PI3K-AKT-mTOR pathway and that ASNS maintained cell adaptation to glutamine depletion through asparagine biosynthesis in KRAS-mutant CRCs. Notably, tumor growth of KRAS-mutant CRC cells was significantly suppressed upon ASNS knockdown. These results indicate that ASNS might be a novel therapeutic target for CRCs with mutated KRAS. To our knowledge, this is the first study to investigate amino acid metabolism associated with KRAS mutation in CRC.
Materials and Methods
Cell Lines and Reagents
All lines were maintained in Dulbecco's modified Eagle medium (DMEM) (glucose 25 mM, glutamine 4 mM) (043-30085, Wako) containing 10% FBS. Media without glutamine were prepared by using glutamine-free DMEM (glucose 25 mM, glutamine 0 mM) (043-32245, Wako) containing 10% FBS. LoVo, RKO, COLO-205, and WiDR cells were supplied from American Type Culture Collection. The identity of cell line was confirmed by short tandem repeat analysis (Takara Bio). HCT116, HKh-2, HKe-3, DLD-1, and DKO-4 cell lines were generated as described previously [21]. All amino acid were obtained from Wako. U0126 was purchased from Calbiochem; LY294002 and rapamycin were from Wako.
Measurement of Metabolites
In cell samples, 1 × 106 cells were seeded in 10-cm dishes in DMEM (25 mM glucose, 4 mM glutamine) containing 10% FBS for 48 hours. Metabolome analysis was conducted by C-SCOPE package of Human Metabolome Technologies, Inc., using capillary electrophoresis (CE) time-of-flight mass spectrometry for cation analysis and CE tandem mass spectrometry for anion analysis as described previously [11].
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis
Total RNAs were extracted and reverse transcription was performed with oligo (dT) primer and Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer's instructions. The resulting cDNA was quantified using StepOnePlus Real-Time PCR System (Applied Biosystems) and THUNDERBIRD SYBR qPCR Mix (TOYOBO). Primer sequences are found in Supplementary Table 1. All transcription levels were normalized to that for ACTB.
Western Blot Analysis
Cells and tissue samples were lysed in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 1% NP40) supplemented with inhibitor cocktails of protease and phosphatase. Primary antibodies were mouse monoclonal anti-KRAS (Santa Cruz Biotechnology), rabbit polyclonal anti-ASNS (Abcam), rabbit monoclonal anti–pohospho-p44/42 kinase (Thr202/Tyr204) (Cell Signaling), rabbit monoclonal anti-p44/42 kinase (Cell Signaling), rabbit monoclonal anti–phospho-Akt (Ser473) (Cell Signaling), rabbit monoclonal anti-Akt (Cell Signaling), rabbit polyclonal anti–phospho-p70 S6 kinase (Thr389) (Cell Signaling), rabbit polyclonal anti-p70 S6 kinase (Cell Signaling), and mouse monoclonal anti–β-actin-peroxidase (Sigma-Aldrich).
RNA Interference
To knock down the endogenous expressions of both KRAS and CS, FlexiTube GeneSolutions (Qiagen) for KRAS (#1: SI03101903, #2: SI03106824) and CS (#1: SI04147997, #2: SI04184796) were employed, and AllStars negative control (NC) small interfering RNA (siRNA) was used for nonsilencing control. The siRNA (10 nM) was transfected with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instruction.
Cell Proliferation Assay
A cell proliferation assay was performed with Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer's instruction. Cells were cultured in 96-well plates at a density of 5000 cells/well for 48 to 72 hours with or without treatment.
Clonogenic Assay
Cells were seeded in 6-well plates at a density of 200 cells/well in DMEM (25 mM glucose, 4 mM glutamine) containing 10% FBS. Ten days later, colonies were fixed in 1% glutaraldehyde and stained with 0.2% crystal violet for 30 minutes. A colony was defined as a cluster of at least 50 cells.
Construction of Recombinant Lentivirus
The following oligonucleotides were used to knock down ASNS expression: shASNS#1-sense, 5′-CCGGATGGTGAAATCTACAACCATACTCGAGTATGGTTGTAGATTTCACCAT-TTTTTG-3′;shASNS#1-antisense, 5′-AATTCAAAAAATGGTGAAATCTACAACCATACTCGAGTATGGTTGTAGATTTCACCAT-3′;shASNS#2-sense, 5′-CCGGTTAGGTGGTCTTTATGCTGTACTCGAGTACAGCATAAAGACCACCTAATTTTTG-3′;shASNS#2-antisense, 5′-AATTCAAAAATTAGGTGGTCTTTATGCTGTACTCGAGTACAGCATAAAGACCACCTAA-3′. Each set of oligonucleotides was annealed and cloned into the AgeI/EcoRI sites of pLKO.1 (Addgene). pLKO.1-TRC control vector (Addgene) was used as control. ASNS cDNA was obtained from HCT116 cells. The ASNS cDNA was cloned into the pLVSIN-CMV (TaKaRa BIO).
Apoptosis Assay
In terminal deoxynucleotidyl transferase-mediated dUTP-FIFC nick end-labeling (TUNEL) assay, cells were cultured under glutamine-free DMEM (25 mM glucose, 0 mM glutamine) containing 10% FBS for 72 hours and then fixed in 4% parafolmaldehyde. The number of TUNEL-positive cells in each of the three slides was counted in five random fields (magnification, ×400) by software BZ-H3C (Keyence) and expressed as a percentage of the total cells counted. The activity of caspase-3 and -7 was measured by using caspase-Glo 3/7 assay (Promega) according to the manufacturer's protocol. Caspase activity was normalized to the cell number counted by CCK-8 cell proliferation assay under the same density and conditions.
Animal Tumor Model
We subcutaneously injected 4-6 × 106 cancer cells into flanks of 4- to 6-week-old female KSN/slc nude mice (JAPAN SLC). Tumor volumes were calculated using the formula (L × W2) × 0.5, where L is length and W is width. At the time of L-asparaginase (L-Asp) and rapamycin treatment, tumors were established in KSN/slc nude mice by subcutaneous injection of 5 × 106 of HCT116. Mice with tumors of 150 to 300 mm3 were randomly divides into 4 groups. L-Asp (Kyowa Hakko Kirin Co., Ltd.) was injected daily intraperitoneally at 1500 U/kg for 15 days [22]. Rapamycin (LC Laboratories) was dosed in 4% ethanol/5% PEG400/5% Tween 80 in water for injection solution and injected every 5 days intraperitoneally at 3 mg/kg [23]. The control group was injected every 5 days with an equal volume of the vehicle. Mice were ethically sacrificed when mean tumor volume reached 2000 mm3. Animal experimental studies were conducted in accordance with our institutional guidelines and approved by the Kyoto University Animal Care Committee.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were stained with anti-rabbit ASNS (Abcam), anti-mouse CD34 (Abcam), or ant-mouse Ki67 (Dako) antibody. Antigen retrieval was achieved with microwave in citrate buffer (pH: 6.0). The Ki67-positive cell numbers were counted in five random fields (×400). For primary CRC tissue, ASNS immunoreactivity score was determined by the sum of distribution and staining intensity, as previously described [24]. ASNS expression was defined as low (total score 0-3) or high (total score 4-7). The distribution was scored based on the positivity rate as “0” (0%-5%), “1” (5%-25%), “2” (25%-50%), “3” (50%-75%), or “4” (>75%). The staining intensity was scored as “0” (no staining), “1” (weakly stained), “2” (moderately stained), or “3” (strongly stained). Two researchers (K.T. and S.I.) independently evaluated all immunohistochemistry samples without prior knowledge of other data. The slides with different evaluations among them were reinterpreted at a conference to reach the consensus.
Patients, Clinicopathological Data
A total of 93 CRC samples were collected from patients who underwent tumor resection and with KRAS status checked at Kyoto University Hospital between April 2009 and September 2013. No patients received preoperative chemotherapy and radiation therapy. This study protocol was approved by the institutional review board of Kyoto University, and patients provided their written consent for data handling.
Statistical Analysis
The statistical significance of differences was determined by Student's t test or χ2 test. All values were expressed as mean ± standard deviation (SD). All analyses were two-sided, and differences with a P value of less than .05 were considered statistically significant in all analyses. Statistical analyses were conducted with the JMP software (version 9, SAS Institute, Inc.).
Results
KRAS Mutation-Related Alterations of Amino Acid Metabolism in Human CRC Cells
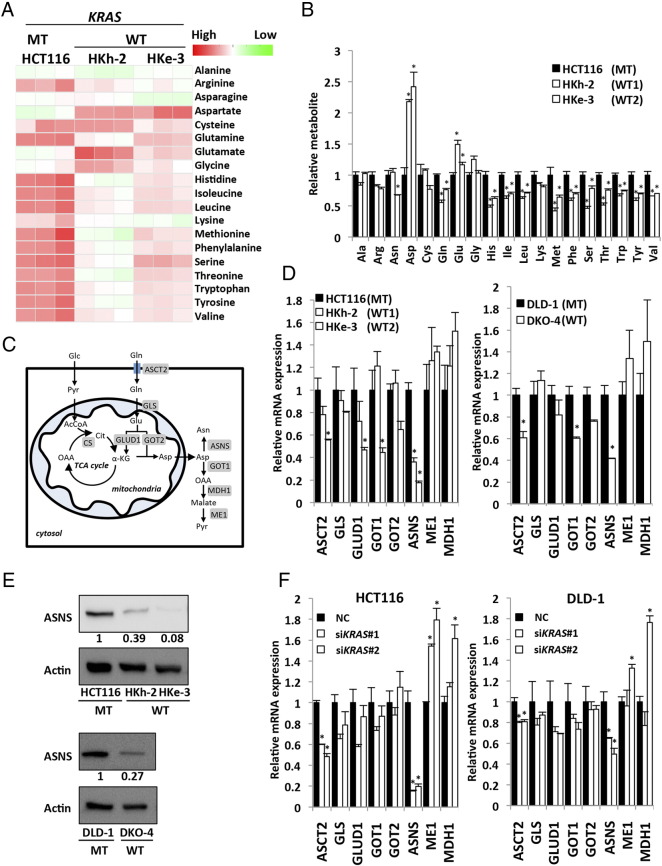
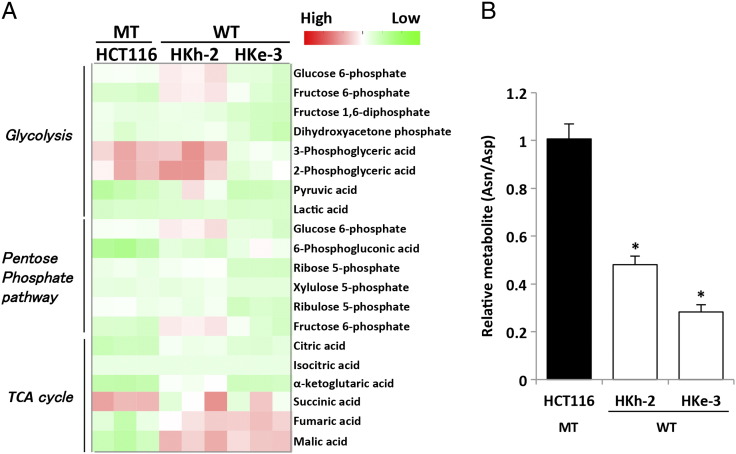
To investigate the specific pathway by which mutated KRAS exhibits metabolic alterations, we used human CRC cell lines with mutated KRAS (HCT116 and DLD-1) and their isogenic lines in which the endogenous mutant KRAS allele was disrupted through targeted homologous recombination [11], [21]. HKh-2 and HKe-3 are two independent HCT116 clones with the mutated KRAS disrupted; similarly, DKO-4 is a DLD-1 clone with the mutated KRAS disrupted. These paired cell lines essentially differ only in the mutational status of KRAS gene, which allowed us to investigate the function of normal or mutant KRAS proteins at physiological, normally regulated levels. In addition, these CRC lines also harbor additional mutations in CTNNB1 and TGFβ-RII genes [25], [26]; therefore, KRAS signaling can be studied in the context of other gene mutations commonly found in CRC. Using these paired lines, we previously reported that mutated KRAS caused higher FDG accumulation by upregulation of glucose uptake through GLUT1 [10], [11], [12]. To further dissect the metabolic alterations caused by mutated KRAS, CE mass spectrometry (CE/MS) metabolomic analysis was performed to comprehensively characterize intracellular metabolites in HCT116 paired lines. This analysis revealed that mutated KRAS induced changes in multiple metabolic pathways such as glycolysis, PPP, TCA, and most significantly the amino acid pathway (Figure 1A and Supplementary Figure S1A). In particular, aspartate level in HCT116 with mutated KRAS was significantly lower compared with that in their isogenic counterparts containing wild-type KRAS allele (HKh-2 and HKe-3). In contrast, asparagine level was significantly higher in HCT116 than in HKe-3 (Figure 1B). The asparagine/aspartate ratio was significantly higher in HCT116 than in HKh-2 and HKe-3 (Supplementary Figure S1B).
Figure 1.
Metabolic alterations caused by KRAS mutations in colorectal cancer cells.
(A) Heat map shows metabolic changes of amino acid in KRAS-mutant CRC cells (HCT116) and their isogenic counterparts containing wild-type KRAS allele (HKh-2 and HKe-3). Metabolome analysis (CE/MS) was performed from triplicates for each condition and normalized to cell number. Cells were maintained in DMEM (25 mM glucose, 4 mM glutamine) containing 10% FBS for 48 hours. WT, KRAS wild type; MT, KRAS mutant. (B) Relative metabolite levels are expressed as fold change compared with metabolite concentration of HCT116. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). Ala, alanine; Arg, arginine; Asn, asparagine; Asp, aspartate; Cys, cysteine; Gln, glutamine; Glu, glutamate; Gly, glycine; His, histidine; Ile, isoleucine; Leu, leucine; Lys, lysine; Met, methionine; Phe, phenylalanine; Ser, serine; Thr, Threonine; Trp, tryptophan; Tyr, tyrosine; Val, valine. (C) Schematic representation of glutamine metabolism. α-KG, α-ketoglutarate; OAA, oxaloacetate; Pyr, pyruvate; Glc, glucose; AcCoA, acetyl-CoA; Cit, citrate. (D) Quantitative RT-PCR showing relative mRNA levels for metabolic enzymes associated with glutamine metabolism. KRAS-mutant cells (HCT116 and DLD-1) and their isogenic counterpart containing wild-type KRAS allele (HKh-2, HKe-3, and DKO-4). Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (E) Western blot photographs of ASNS and β-actin (Actin) (top) and quantified data for ASNS (bottom) (normalized with β-actin). (F) Quantitative RT-PCR showing relative mRNA levels for metabolic enzymes associated with glutamine metabolism. HCT116 cells (left) and DLD-1 cells (right) were treated separately with two independent siRNA constructs (#1 and #2) targeting KRAS and NC siRNA. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05).
Supplementary Figure S1.
Metabolic alterations caused by KRAS mutations in colorectal cancer cells. (A) Heat map shows metabolic changes in KRAS-mutant CRC cells (HCT116) and their isogenic counterparts containing wild-type KRAS allele (HKh-2 and HKe-3). Metabolome analysis (CE/MS) was performed from triplicates for each condition and normalized to cell number. Cells was maintained in DMEM (25 mM glucose, 4 mM glutamine) containing 10% FBS for 48 hours. WT, KRAS wild type; MT, KRAS mutant. (B) Intracellular asparagine (Asn)/aspartate (Asp) ratio in CRC cell lines was quantified by metabolome analysis.
Upregulation of Asparagine Synthetase in KRAS-Mutant CRCs Through the PI3K-AKT-mTOR Pathway
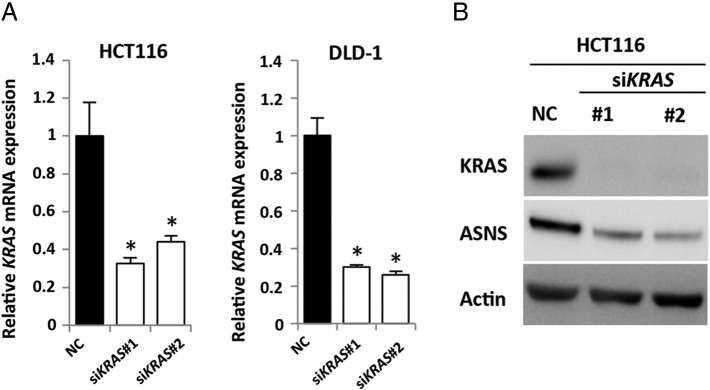
Recently, it has been reported that KRAS mutation in PDCA cells regulates the glutamine metabolism through its conversion to aspartate, thereby supporting tumor growth by maintaining the cellular redox balance [19]. Glutamine is transported into the cytoplasm by neutral amino acid transporter (ASCT2) and is subsequently converted to glutamate by glutaminase (GLS) at mitochondria. Thereafter, glutamate can be converted to α-ketoglutarate by GLUD1 to replenish the TCA cycle by GLUD1 or converted to nonessential amino acids such as aspartate and alanine by transaminases. In PDCA, glutamic-oxaloacetic transaminase 2 (GOT2) was identified as the transaminase associated with mutated KRAS at mitochondria [19]. Glutamine-derived aspartate is transported into the cytoplasm where it can be converted to asparagine by ASNS or to pyruvate by GOT1, malic enzyme 1, and malate dehydrogenase 1 (Figure 1C). A recent report demonstrated that mutated KRAS controlled the reprograming of glutamine metabolism by decreasing GLUD1 and increasing GOT1 in PDCA [19]. To identify the metabolic enzymes associated with the reprograming of glutamine metabolism in CRC, we first screened the expression of the above-mentioned enzymes by quantitative RT-PCR analysis. Notably, ASNS mRNA expression in the parental KRAS-mutant cells (HCT116 and DLD-1) was about 2.5- to 5-fold higher than in their isogenic wild-type KRAS cells (Figure 1D). By Western blotting, we also confirmed that ASNS protein level was about 2.5- to 10-fold higher in the parental KRAS-mutant cells than in the corresponding isogenic wild-type KRAS cells (Figure 1E). To further validate the relationship between ASNS and KRAS, we performed RNA interference experiments. We introduced two kinds of different siRNA constructs targeting KRAS (referred to as siKRAS#1 and #2) into HCT116 and DLD1 cells (Supplementary Figure S2A and B) and verified that both siKRAS constructs dramatically decreased ASNS mRNA expression of ASNS but not GLUD1 or GOT1 (Figure 1F).
Supplementary Figure S2.
Effects of KRAS knockdown in CRC cell lines. (A) Quantitative RT-PCR showing relative mRNA levels for KRAS. HCT116 cells (left) and DLD-1 cells (right) were treated separately with two independent siRNA constructs (#1 and #2) targeting KRAS and NC siRNA. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (B) Western blot photographs of KRAS, ASNS, and β-actin (Actin). HCT116 cells were treated separately with two independent siRNA constructs (#1 and #2) targeting KRAS and NC siRNA.
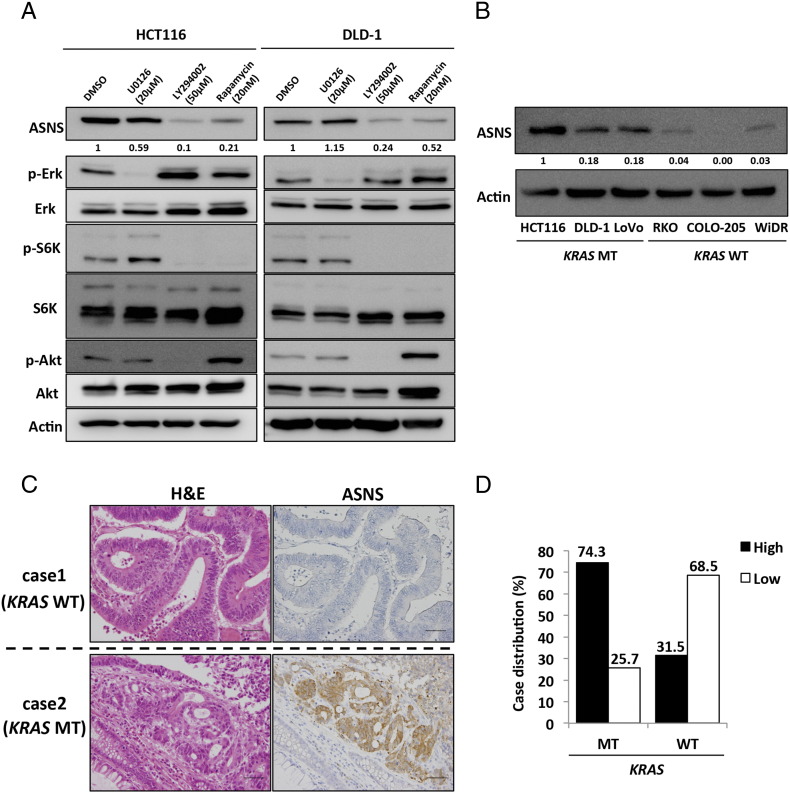
KRAS protein activates its downstream signaling pathways, such as the Raf/MEK/ERK and PI3K/Akt pathways, leading to malignant transformation. We used a specific inhibitor of each pathway to examine which pathway regulates the induction of ASNS expression mediated by mutated KRAS. HCT116 and DLD-1 cells were treated with a MEK inhibitor (U0126, 20 μM), a PI3K inhibitor (LY294002, 50 μM), or an mTOR inhibitor (rapamycin, 20 nM). A Western blot analysis showed that ASNS expression was dramatically inhibited by LY294002 and rapamycin but not U0126 (Figure 2A), suggesting that ASNS expression is correlated with the PI3K/AKT/mTOR pathway in KRAS-mutant cells (HCT116 and DLD-1).
Figure 2.
ASNS expression in CRC.
(A) Western blot analyses of HCT116 (left) and DLD-1 (right) treated with dimethyl sulfoxide (DMSO), 20 μM U0126 (MEK inhibitor), 50 μM LY294002 (PI3K inhibitor), or 20 nM rapamycin (mTOR inhibitor) for 48 hours. Protein levels were normalized to β-actin (Actin). Densitometry values were expressed as fold change compared with DMSO treating cells. (B) Western blot analysis of ASNS expression in several human CRC cell lines. Densitometry values were expressed as fold change compared with HCT116. (C) H&E and immunohistochemical staining for ASNS in primary CRCs. Upper panels show serial sections of a KRAS-mutant CRC case, whereas lower panels show those of a wild-type KRAS one. Scale bar, 50 μm. (D) Relationship between KRAS mutational status and ASNS expression in 93 primary CRC samples.
Based on these findings, we hypothesized that ASNS expression in CRC is regulated by mutated KRAS. We therefore determined the expression level of ASNS in several human CRC cell lines. HCT116, DLD-1, and LoVo cells bear a mutant KRAS allele at codon 13 (G13D), whereas RKO, COLO-205, and WiDr cells bear the wild-type KRAS. A Western blot analysis showed that KRAS-mutant cells constitutively expressed high level of ASNS, whereas wild-type KRAS cells exhibited little to no ASNS expression (Figure 2B). To further investigate the relationship between KRAS status and ASNS expression, we next examined clinical specimens of human primary CRC by immunohistochemistry. Representative cases of CRCs bearing mutated and wild-type KRAS are shown in Figure 2C. Of a total of 93 primary CRC patients, 50% (46 of 93) were high for ASNS, whereas 50% (47 of 93) were low (Table 1). Patients were classified into two groups based on their KRAS mutational status: patients with mutated KRAS (n = 39) and those with wild-type KRAS (n = 54). ASNS expression was high in 74% (29 of 39) of patients with mutated KRAS, whereas it was low in 31% (17 of 54) of those with wild-type KRAS, with a significant correlation between mutated KRAS and ASNS expression (odds ratio, 6.31; P < .001, Figure 2D). ASNS expression was not correlated with age, sex, tumor location, tumor size, T-category, N-category, and M-category (Table 1). Taken together, KRAS status is one of the key regulators of ASNS expression in CRC.
Table 1.
Univariate Analysis of Factors Associated With Expressions of ASNS
| ASNS |
|||
|---|---|---|---|
| High (n = 46) | Low (n = 47) | P | |
| Mean, age ± SD (y) | 69.5 ± 10.7 | 69.5 ± 10.8 | .63 |
| Sex | |||
| Male | 28 | 27 | .84 |
| Female | 18 | 20 | |
| Location | |||
| Colon | 32 | 33 | 1 |
| Rectum | 14 | 14 | |
| Mean tumor size (mm) | .26 | ||
| ≥50 | 24 | 19 | |
| <50 | 22 | 28 | |
| UICC-TMN stage | |||
| I/II | 20 | 27 | .22 |
| III/IV | 26 | 20 | |
| T-category | |||
| Negative | 12 | 12 | 1 |
| Positive | 34 | 35 | |
| M-category | |||
| Negative | 39 | 39 | 1 |
| Positive | 7 | 8 | |
| N-category | |||
| Negative | 22 | 29 | .21 |
| Positive | 24 | 18 | |
| KRAS | |||
| Wild type | 17 | 37 | <.001 |
| Mutated | 29 | 10 | |
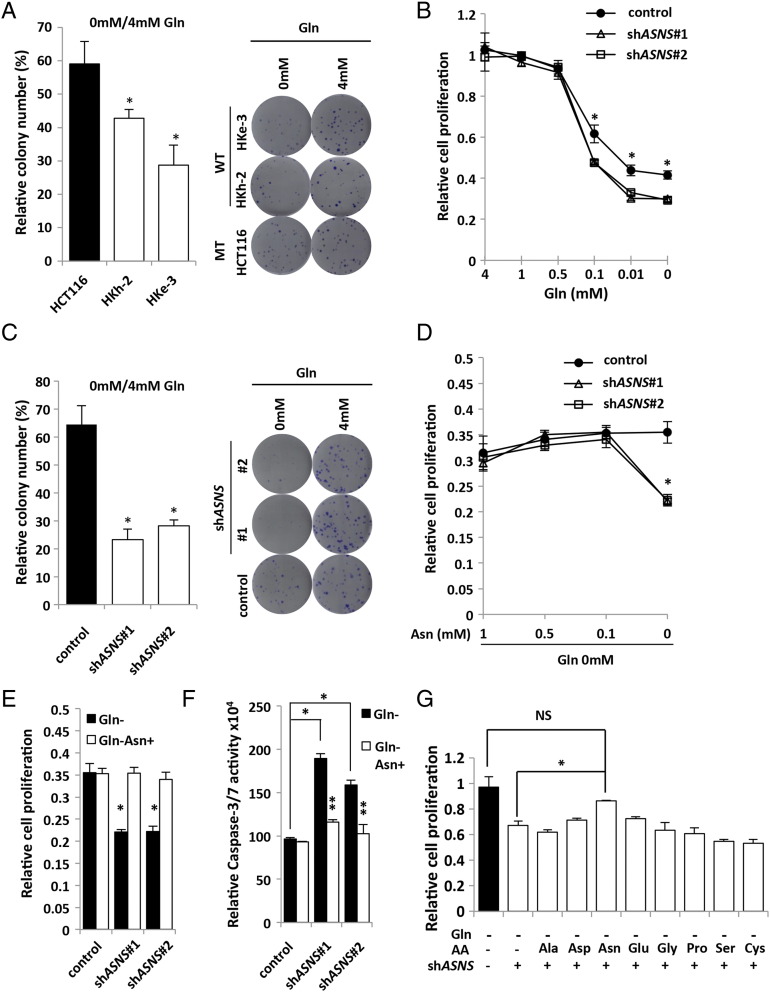
The Role of ASNS in Cell Adaptation to Glutamine Depletion in KRAS-Mutant CRC Cells
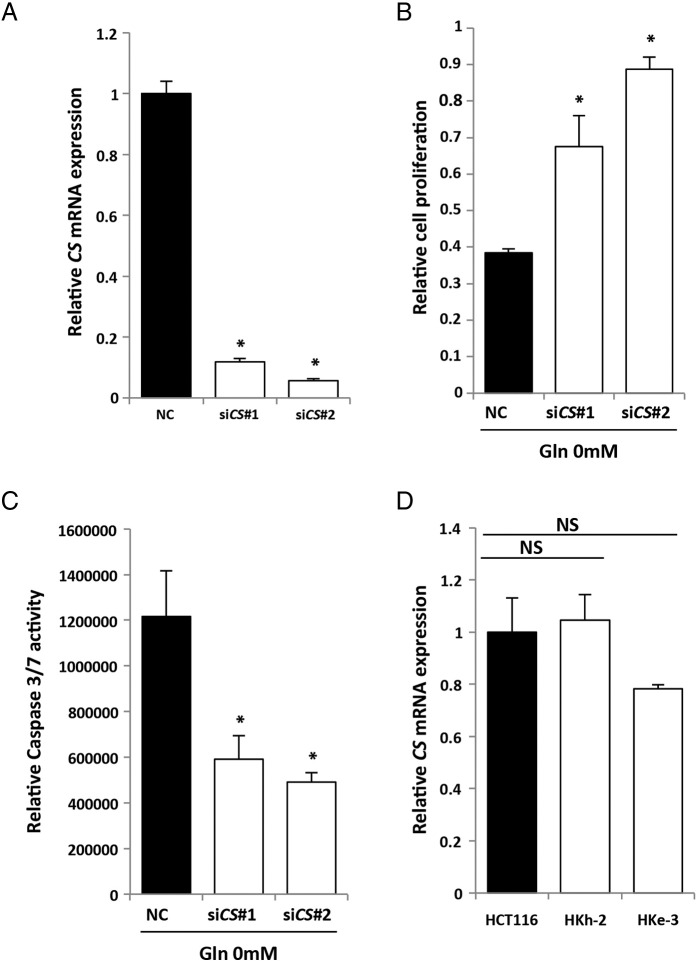
ASNS is the enzyme that synthesizes de novo asparagine from aspartate using glutamine as a nitrogen source. In human glioma and neuroblastoma, it was recently reported that asparagine was essential to suppress apoptosis induced by glutamine withdrawal without restoring the levels of other nonessential amino acids or TCA cycle intermediates and that ASNS knockdown led to cell death even in the presence of glutamine [20]. To evaluate whether KRAS mutational status of CRC cells could affect their addiction to glutamine, we tested the clonogenic assay. In the absence of glutamine, colony number of KRAS-mutant cells (HCT116) was significantly higher than the isogenic wild-type KRAS cells (HKh-2 and HKe-3) (Figure 3A), indicating that KRAS-mutant CRC cells were more resistant to glutamine deprivation than wild-type KRAS CRC cells. Citrate synthase (CS) is a TCA cycle enzyme that catalyzes the formation of citrate from oxaloacetate and acetyl-CoA. CS siRNA was identified to be a suppressor of glutamine withdrawal–induced apoptosis by reducing TCA cycle activity and redirecting oxaloacetate for synthesis of aspartate and asparagine [20]. Consistent with the findings in human glioma and neuroblastoma, we found that CS knockdown using siRNA prevented glutamine withdrawal–induced apoptosis and suppression of cell proliferation in HCT116 cells (Supplementary Figure S3, A–C), although CS expression level was not correlated with KRAS mutation (Supplementary Figure S3D). These data further support the critical role of this pathway in glutamine metabolism and tumor growth.
Figure 3.
ASNS maintains cell adaptation to glutamine depletion in KRAS-mutant CRC cells.
(A) Clonogenic assay with HCT116 and their isogenic wild-type KRAS cells (HKh-2 and HKe-3). Cells were maintained under 4 mM or 0 mM glutamine (Gln) condition containing 10% FBS for 10 days. Each colony number in 0 mM Gln was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (B) Cell proliferation measured by CCK-8 assay. HCT116 cells transfected with control or two independent shASNS vectors were maintained under various Gln concentration containing 10% FBS for 48 hours. Viability in each condition was normalized to that in 4 mM Gln. Student's t test; *P < .05. (C) Clonogenic assay with HCT116 transfected with control or two independent shASNS vectors. Cells were maintained under 4 or 0 mM Gln condition containing 10% FBS for 10 days. Each colony number in 0 mM Gln was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (D) Cell proliferation measured by CCK-8 assay. HCT116 transfected with control or two independent shASNS vectors. Cells were maintained under various concentrations of Asn and 0 mM Gln containing 10% FBS for 48 hours. Viability in each condition was normalized to that in 4 mM Gln. Student's t test; *P < .05. (E) Cell proliferation measured by CCK-8 assay. HCT116 cells transfected with control or two independent shASNS vectors were maintained under either 0 mM Gln or 0 mM Gln + 0.1 mM Asn containing 10% FBS for 48 hours. Viability in each condition was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (F) Caspase 3/7 activities. HCT116 transfected with control or two independent shASNS vectors were maintained under either 0 mM Gln or 0 mM Gln + 1 mM Asn containing 10% FBS for 48 hours. Caspase 3/7 activity was normalized to the cell viability measured by CCK-8 assay under the same density and conditions. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, control vector versus shASNS vectors; **P < .05, 0 mM Gln versus 0 mM Gln+ 1 mM Asn). (G) Cell proliferation measured by CCK-8 assay. HCT116 cells transfected with control or shASNS vectors were maintained under 0 mM Gln plus each amino acid (0.5 mM) containing 10% FBS for 48 hours. Viability in each condition was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, NS: not significant).
Supplementary Figure S3.
CS and CRC cells. (A) Quantitative RT-PCR showing relative mRNA levels for CS. HCT116 cells were treated separately with two independent siRNA constructs (#1 and #2) targeting CS and NC siRNA. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (B) Cell proliferation measured by CCK-8 assay. HCT116 transfected with control or two independent siCS were maintained under 0 mM Gln containing 10% FBS for 48 hours. Viability in each condition was normalized to that in 4 mM Gln. Student's t test; *P < .05. (C) Caspase 3/7 activities. HCT116 transfected with control or two independent siCS were maintained under 0 mM Gln containing 10% FBS for 48 hours. Caspase 3/7 activity was normalized to the cell viability measured by CCK-8 assay under the same density and conditions. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (D) Quantitative RT-PCR showing relative mRNA levels for CS in KRAS-mutant CRC cells (HCT116) and their isogenic counterparts containing wild-type KRAS allele (HKh-2 and HKe-3). Mean; bars, ± SD (Student's t test; NS: not significant).
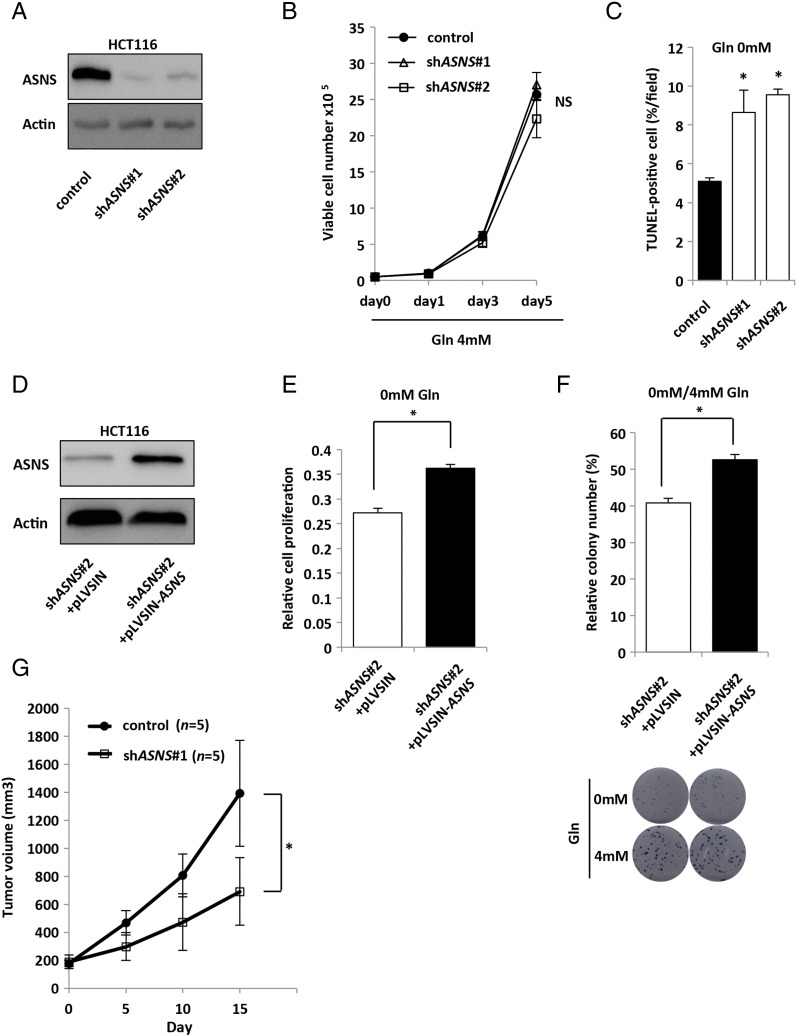
To further investigate the role of ASNS, two lentiviral shRNA constructs targeting ASNS (referred to as shASNS#1 and shASNS#2) were introduced into HCT116 cells (Supplementary Figure S4A). Under the condition of glutamine depletion, both shASNS constructs significantly decreased cell proliferation compared with the control in a dose-dependent manner (Figure 3B), although cell proliferation was not affected under sufficient amount of glutamine (4 mM Gln) (Supplementary Figure S4B). Similarly, under the glutamine-free condition, the colony number of shASNS transfectants was significantly lower than that of the control cells in the clonogenic assay (Figure 3C). We also found that the number of apoptotic cells was significantly higher in shASNS transfectants than the control cells (Supplementary Figure S4C). To test whether these effects of shRNAs were specific, we further performed rescue experiments. Namely, the ASNS expression plasmid (pLVSIN-ASNS) was transfected into shASNS#2 cells in which the targeted sequence was located at the 3′-untranslated region (3′UTR) (Supplementary Figure S4D). Both the proliferation rate and clonogenic growth of shASNS transfectants under the glutamine-free condition were significantly rescued by ASNS overexpression (Supplementary Figure S4, E and F). To evaluate whether the growth-inhibitory effects of shASNS constructs were recovered by exogenous addition of asparagine (the ASNS product), we next tested the effect of asparagine addiction to the medium and found that 0.1 mM asparagine was sufficient to recover the growth inhibition of shASNS transfectants in the cell proliferation (Figure 3, D and E) and clonogenic assays (Supplementary Figure S5A). Consistently, asparagine addition significantly suppressed the apoptosis of shASNS transfectants under the glutamine-free condition (Figure 3F). Furthermore, we found that the significant growth recovery of shASNS transfectants under the glutamine-free condition was observed only upon asparagine addition but not other amino acids (Figure 3G), indicating the specificity of asparagine-mediated rescue of cell survival. Together, these data indicate that ASNS upregulation caused by KRAS mutation maintains cell adaptation to glutamine depletion through asparagine biosynthesis.
Supplementary Figure S4.
The effects of ASNS in CRC cells. (A) Western blot photographs of ASNS and β-actin (Actin). HCT116 cells were treated separately with two independent shRNA constructs (#1 and #2) targeting ASNS and control siRNA. (B) Cell proliferation rates of HCT116 transfected with control or two independent shASNS vectors under 4 mM glutamine (Gln) containing 10% FBS. Mean; bars, ± SD (Student's t test; NS: not significant). (C) TUNEL assay of HCT116 transfected with control or two independent shASNS vectors under 0 mM Gln containing 10% FBS. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (D) Western blot photographs of ASNS and β-actin (Actin). ASNS expression vector (pLVSIN-ASNS) or control vector (pLVSIN) was transfected into shASNS#2 cells in which the targeted sequence was located at 3′UTR. (E) Cell proliferation measured under 0 mM Gln containing 10% FBS for 48 hours. ASNS expression vector (pLVSIN-ASNS) or control vector (pLVSIN) was transfected into shASNS#2 cells in which the targeted sequence was located at 3′UTR. Viability in each condition was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (F) ASNS expression vector (pLVSIN-ASNS) or control vector (pLVSIN) was transfected into shASNS#2 cells in which the targeted sequence was located at 3′UTR. Cells were maintained under 4 mM or 0 mM Gln condition containing 10% FBS for 10 days. Each colony number in 0 mM Gln was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (G) Xenograft size of HCT116 transfected with control or shASNS#1 vector. HCT116 transfected subcutaneously injected into the flank of 4- to 6-week-old female nude mice. Tumor volume was measured every 5 days from the start day when all tumors had reached 150 to 200 mm3 in size (each group, n = 5). Student's t test; *P < .05.
Supplementary Figure S5.
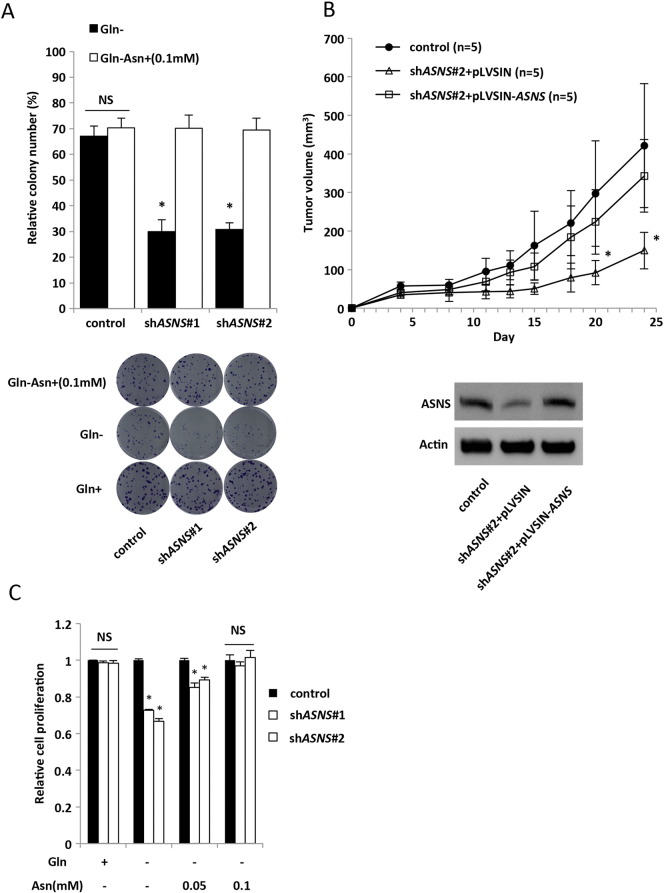
ASNS and Asn. (A) Clonogenic assay with HCT116 transfected with control or two independent shASNS vectors. Cells were maintained under 4 mM glutamine (Gln), 0 mM Gln, or 0 mM Gln + 0.1 mM Asn containing 10% FBS for 10 days. Each colony number was normalized to that in 4 mM Gln. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (B) Xenograft size of HCT116 cells transfected with control, shASNS#2 + pLVSIN, or shASNS#2 + pLVSIN-ASNS. ASNS expression vector (pLVSIN-ASNS) was transfected into shASNS#2 cells in which the targeted sequence was located at 3′UTR. A total of 6 × 106 cells were subcutaneously injected into the flank of 4-week-old female nude mice, and then tumor volumes were measured 2 times per week (each group, n = 5). Xenograft size of HCT116 cells transfected with shASNS#2 + pLVSIN was significantly smaller than that of shASNS#2 + pLVSIN-ASNS. Mean; bars, ± SD (Student's t test; *P < .05 compared with control or shASNS#2 + pLVSIN-ASNS). (C) Cell proliferation measured by CCK-8 assay. HCT116 transfected with control or two independent shASNS vectors. Cells were maintained under 4 mM Gln, 0 mM Gln, or 0 mM Gln + various concentrations of Asn containing 10% FBS for 48 hours. Viability in each condition was normalized to control. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, NS: not significant).
Suppression of Tumor Growth In Vivo of KRAS-Mutant CRC Cells by ASNS Inhibition
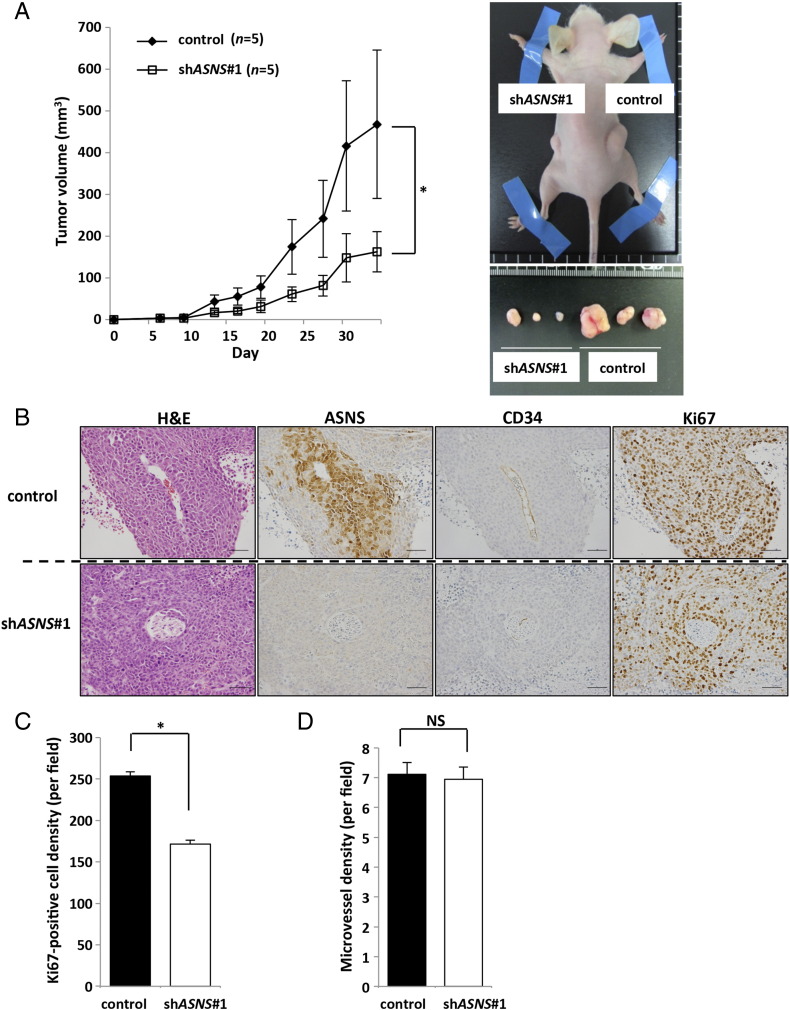
As further investigation of the importance of ASNS in KRAS-mutant CRC cells, we assessed the ability of CRC cells to grow as xenografts in vivo. Consistent with results obtained from the in vitro studies, shASNS#1 construct significantly reduced tumor growth of HCT116 cells compared with the control cells (Figure 4A and Supplementary Figure S4G). Immunohistochemical analysis indicated that the number of Ki67-positive tumor cells was significantly lower in shASNS#1 transfectant tumors than in the control tumors, especially in the location distant from the CD34-positive tumor blood vessels (Figure 4, B and C). We also measured the intratumoral vessel density and found no difference between shASNS#1 transfectant tumors and the control (Figure 4D). Furthermore, we also performed rescue experiments in vivo and found that the reduced tumor growth of shASNS#2 transfectant was significantly recovered by ASNS overexpression (Supplementary Figure S5B).
Figure 4.
ASNS knockdown inhibits tumor growth in vivo.
(A) Xenograft size of HCT116 cells transfected with control or shASNS vector. A total of 4 × 106 cells were subcutaneously injected into the flank of 4- to 6-week-old female nude mice, and then tumor volumes were measured 2 times per week (each group, n = 5). Representative images of tumors are shown. (B) H&E and immumohistochemical staining for ASNS, CD34, and Ki67 in xenograft tumors. Upper panels show serial sections of control tumors, whereas lower panels show those of shASNS-transfected tumors. Scale bar, 50 μm. (C) Ki67-positive cell density. Cell numbers were counted in five random fields (each group, n = 3). Mean; bars, ± SD (Student's t test; *P < .05). (D) Microvessel density. CD34-positive cells were counted in five random fields (each group, n = 3). Mean; bars, ± SD (Student's t test; NS: not significant).
Combination Treatment of L-Asp Plus Rapamycin
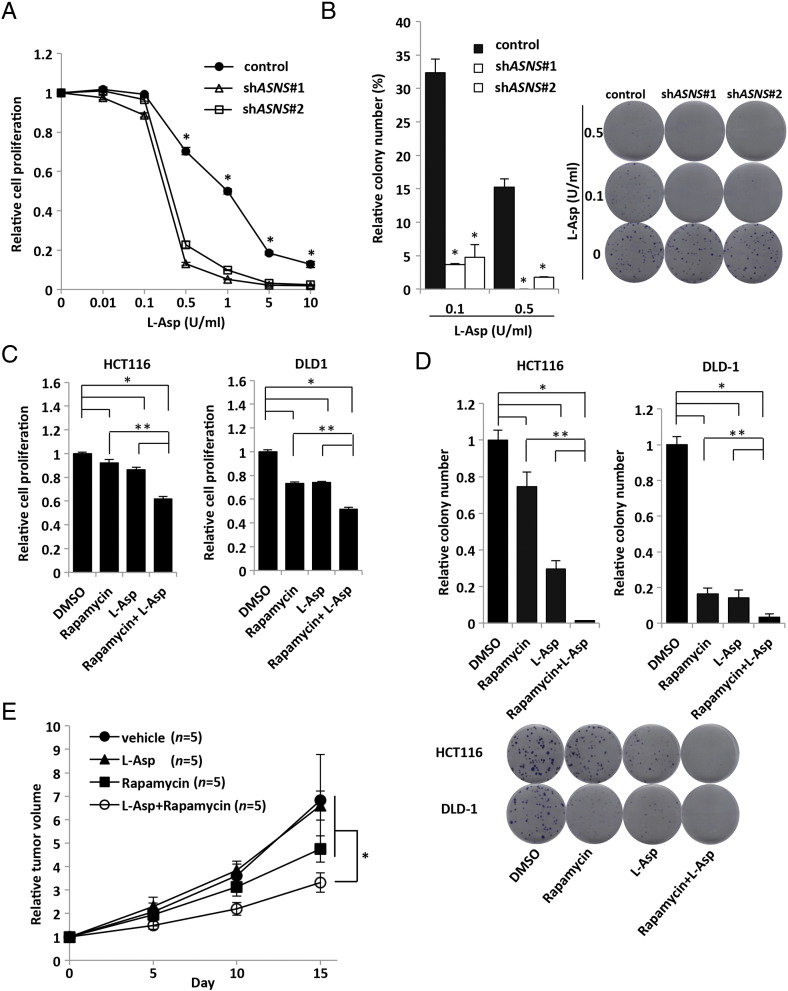
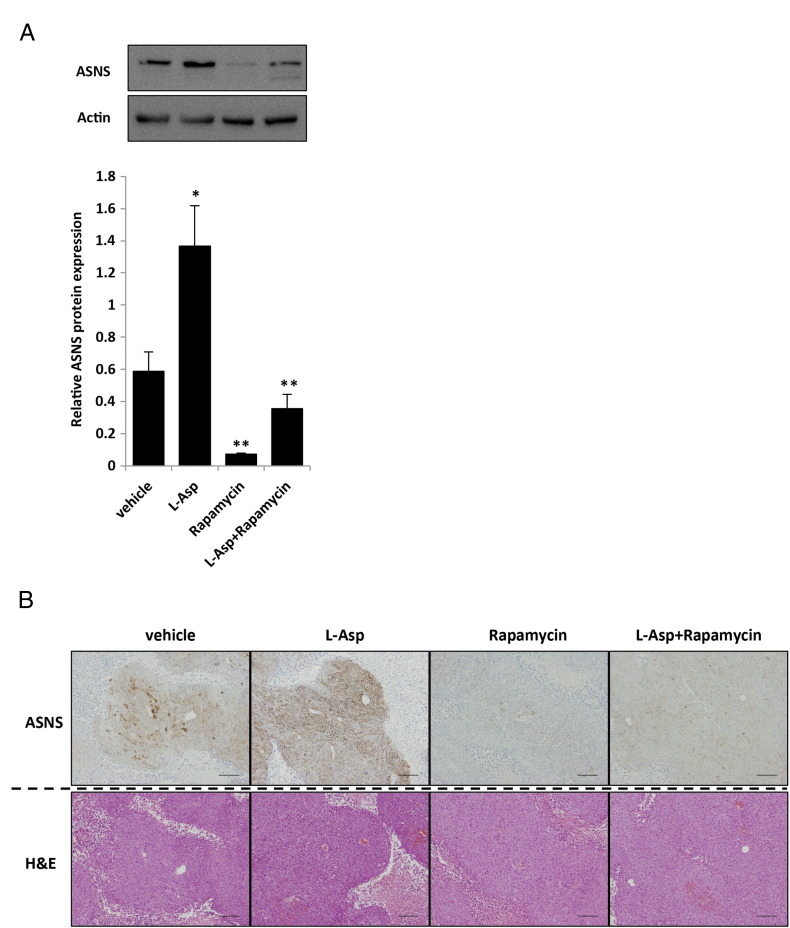
Given the importance of glutamine metabolism in the maintenance of tumor growth, we speculated that KRAS-mutant CRC cells might be sensitive to the inhibition of anabolic glutamine metabolism. Therefore, we determined whether ASNS inhibition combined with depletion of plasma glutamine and asparagine could be used as a novel therapeutic strategy to treat KRAS-mutant CRCs. L-Asp, an FDA-approved drug widely used to treat acute lymphoblastic leukemia (ALL), has activities of both asparaginase and GLS [27], [28]. The inhibitory effect of L-Asp on the cell proliferation and clonogenic growth in vitro was significantly higher in shASNS transfectants than in the control cells (Figure 5, A and B). A potent ASNS inhibitor capable of inhibiting the intracellular asparagine biosynthesis in vivo is yet to be developed [20], [29], [30]. We found that the upregulation of ASNS expression mediated by the mutated KRAS was dramatically suppressed by rapamycin (Figure 2A), and so we examined whether rapamycin could synergize with L-Asp in the clonogenic assay. In both the clonogenic and cell proliferation assays, the combination of L-Asp plus rapamycin suppressed cell growth of KRAS-mutant CRCs (HCT116 and DLD-1) more prominently than L-Asp or rapamycin alone did (Figure 5, C and D). We next assessed the sensitivity of HCT116 xenografts to the combination of L-Asp plus rapamycin in vivo. Mice with tumors of 150 to 300 mm3 were randomly divided into 4 groups and treated with vehicle, L-Asp alone (1500 U/kg), rapamycin alone (3 mg/kg), or the combination of both drugs. Consistent with the in vitro results, tumor growth of xenografts was strongly suppressed by the combination of L-Asp plus rapamycin, although treatments using each single drug showed no or marginal effect (Figure 5E). We examined ASNS expression in vivo of each xenograft and confirmed that rapamycin significantly suppressed ASNS expression, whereas L-Asp upregulated its expression (Supplementary Figure S6, A and B). Taken together, these results indicate that the combination therapy using L-Asp plus ASNS inhibitor could be an effective strategy to treat KRAS-mutant CRC by impairing the glutamine metabolism.
Figure 5.
Inhibition of ASNS suppresses in vivo tumor growth of KRAS-mutant CRC cells.
(A) Cell proliferation measured by CCK-8 assay. HCT116 cells transfected with control or two independent shASNS vectors were treated with various concentrations of L-Asp for 72 hours. Viability in each condition was normalized to the control. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (B) Clonogenic assay with HCT116 transfected with control or two independent shASNS vectors. Cells were treated by various concentrations of L-Asp for 10 days. Each colony number was normalized to that under no treatment of L-Asp. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05). (C) Cell proliferation measured by CCK-8 assay. HCT116 and DLD-1 cells were treated by DMSO, rapamycin (20 nM), L-Asp (0.25 U/ml), or rapamycin (20 nM) + L-Asp (0.25 U/ml) for 48 hours. Viability in each condition was normalized to that of DMSO treatment group. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, compared with DMSO treatment group; **P < .05, compared with combination treatment group [rapamycin + L-Asp]). (D) Clonogenic assay. HCT116 and DLD-1 cells were treated by DMSO, rapamycin (20 nM), L-Asp (0.1 U/ml), or rapamycin (20 nM) + L-Asp (0.1 U/ml). Each colony number was normalized to that of DMSO treatment group. Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, compared with DMSO treatment group; **P < .05, compared with combination treatment group [rapamycin + L-Asp)]. (E) Xenograft size of HCT116 cells treated with vehicle, L-Asp, rapamycin, or L-Asp + rapamycin. HCT116 subcutaneously injected into the flank of 4- to 6-week-old female nude mice. Tumors of 150 to 300 mm3 were randomly divides into 4 groups: vehicle, L-Asp alone (1500 U/kg), rapamycin alone (3 mg/kg), and combination of both drugs groups. We injected L-Asp daily and rapamycin every 5 days intraperitoneally for 15 days. Relative tumor volume was calculated based on the tumor volume at the start day of each treatment. Each group, n = 5. Mean; bars, ± SD, Student's t test; *P < .05, compared with combination treatment group (rapamycin + L-Asp).
Supplementary Figure S6.
Combination of L-Asp plus rapamycin markedly suppresses tumor growth. (A) Western blot analysis of ASNS expression in the xenografts treated by each drug, and quantified data for ASNS (bottom) (normalized with β-actin). Mean; bars, ± SD, n = 3 (Student's t test; *P < .05, control vehicle versus L-Asp treatment; **P < .05, control vehicle versus rapamycin treatment or L-Asp and rapamycin combination treatment). (B) Hematoxylin and eosin (H&E) and immunohistochemical staining for ASNS with each treated xenograft. Scale bar, 100 μm.
Discussion
Metabolic alterations have recently emerged as one of the cancer hallmarks [4], [5], [6], [7]. Major tumor suppressors and oncogenes have intimate connections with the metabolic pathways. KRAS mutations occur in a variety of human malignancies, most frequently in PDCA, non–small cell lung cancer, and CRC. Recent studies indicate that the role of KRAS in cancer metabolism is complex [31], [32]. In an inducible KRASG12D-driven PDCA mouse model, KRASG12D played a vital role in controlling tumor metabolism by increasing glucose uptake, which resulted in activation of protein glycosylation and ribose biogenesis through the HBP and nonoxidative PPP, respectively [8]. Glucose deprivation can increase the mutation rate of KRAS in CRC, which facilitates glucose uptake through the induction of GLUT1 [9]. Moreover, in KRAS-mutant CRC cell lines, vitamin C treatment was recently shown to induce a substantial increase in endogenous reactive oxygen species mediated by increased uptake of the oxidized vitamin C through GLUT1, which inhibited glycolysis at GAPDH to result in an energy crisis and ultimately cell death [33]. We previously reported that glucose uptake, as measured by FDG positron emission tomography scans, was associated with KRAS mutations and GLUT1 expression in primary and metastatic CRCs [10], [12] and that KRAS-mutant CRC cells increased FDG accumulation by upregulating GLUT1 and HIF-1α under hypoxic conditions [11]. Tumorigenesis induced by mutated KRAS was also shown to be dependent on the function of mitochondria to generate reactive oxygen species, particularly through elevated glutamine metabolism [34]. In the present study, using comprehensive CE/MS metabolomics analysis, we found that mutated KRAS significantly altered the amino acid metabolism of CRC cells (Figure 1A and Supplementary Figure S1A). Most importantly, mutated KRAS markedly decreased the aspartate level and increased the asparagine level (Figure 1B and Supplementary Figure S1B). In addition, we demonstrated that the expression of ASNS, the enzyme that synthesizes asparagine from aspartate, was upregulated by mutated KRAS in CRC using multiple human CRC cell lines and clinical specimens of human primary CRC (Figure 2, B–D) and that ASNS expression was regulated by a KRAS-activated pathway, specifically the PI3K-AKT-mTOR pathway (Figure 2A). ASNS was reported to be activated by mutated p53, protein limitation, and tumor microenvironment stress [35], [36], [37]. ASNS was reported to be associated with the sensitivity of L-Asp in the treatment of ALL [35], [38]. In addition, ASNS was also reported to be associated with the sensitivity of cisplatin and with clinical prognosis in some types of solid cancer [20], [24], [39], [40], [41], [42]. To our knowledge, this is the first study to investigate the relationship between CRC and ASNS.
In addition to the well-established role of an altered glucose metabolism in cancer, recent studies have also elucidated the role of amino acid metabolism, especially glutamine, in cell proliferation and maintenance [13], [14]. Although the molecular mechanism of glutamine addiction in cancer cells is unknown, several oncogenic mutations or alterations have been linked to glutamine dependency in cancers. For example, Myc is able to increase glutamine metabolism by upregulating GLS expression, which leads to glutamine entry into the TCA cycle as α-ketoglutarate [43]. Moreover, recent studies have revealed that PDCA, glioma, and neuroblastoma metabolize glutamine in a manner that is different from canonical GLUD1 pathway (conversion of glutamine-derived glutamate into α-ketoglutarate by GLUD1 to fuel the TCA cycle) [19], [20]. In PDCA, mutated KRAS contributes to glutamine dependency by using glutamine-derived aspartate to produce oxaloacetate via GOT1 in the cytoplasm. Oxaloacetate is subsequently converted into malate and then to pyruvate to maintain a high NADPH/NADP+ ratio for redox homeostasis [19]. In glioma and neuroblastoma, CS RNAi knockdown prevents apoptosis induced by glutamine withdrawal by reducing TCA cycle activity and diverting oxaloacetate into aspartate and asparagine, which indicates that maintaining TCA cycle is not the primary mechanism of apoptosis suppression by glutamine metabolism. Glutamine-dependent asparagine synthesis by ASNS is required to suppress apoptosis induced by glutamine withdrawal [20]. However, the role of the glutamine metabolism in CRCs has not been fully elucidated. In the present study, we demonstrated that KRAS-mutant CRC cells could become adaptive to glutamine depletion through asparagine biosynthesis by ASNS and that addition of exogenous asparagine rescued the growth inhibition and cell death under the glutamine-free condition (Figure 3). It was recently reported that asparagine addition to glutamine-deprived cells altered the transcriptional response, thereby suppressing the induction of apoptotic regulators of the unfolded protein response effectors CHOP and XBP1 [20]. Moreover, a recent report has indicated that intracellular asparagine promotes cancer cell proliferation through uptake of amino acids (especially serine, arginine, and histidine) which are involved in protein and nucleotide synthesis [44]. Taken together, these results suggest important roles of asparagine in controlling protein synthesis and turnover in CRC. We observed that mutated KRAS did not alter the expression of GLUD1 and GOT1 in CRC (Figure 1, D and F), although mutated KRAS was reported to cause a decrease of GLUD1 and an increase of GOT1 in PDCA [19], indicating that the multifaceted roles of KRAS in metabolism are cell type dependent.
The importance of asparagine to cell survival has already been established through the use of L-Asp as a therapy for ALL. Leukemic cells lack the constitutive expression of ASNS and the ability to synthesize asparagine from glutamine and thus are dependent on exogenous asparagine to maintain their viability. In contrast, most solid tumors express ASNS and synthesize asparagine from glutamine [37], and L-Asp has not been proven as an effective therapy. The effects of ASNS in tumorigenesis also seem to be cell type dependent. It was recently reported that ASNS silencing produced the strongest inhibitory effect on sarcoma growth in a functional genomic shRNA-based screening of mouse sarcomas generated by oncogenic KRAS and disruption of Cdkn2a[22]. ASNS silencing in sarcoma cell lines reduced the percentage of S phase cells and impeded new polypeptide synthesis. The effects of ASNS silencing in sarcoma cells were reversed by exogenous supplementation of asparagine. Moreover, ASNS inhibition significantly slowed mouse sarcoma growth in vivo only when combined with asparaginase, implicating asparagine-promoted cellular adaptation to metabolic stress such as glutamine depletion. In the present study, ASNS inhibition significantly suppressed tumor growth in vivo of KRAS-mutant CRC cells (HCT116) (Figure 4A and Supplementary Figure S4G). Normal serum asparagine levels of mice and humans were reported to be 0.052 mM and 0.046 mM, respectively [22], [45]. In shASNS transfectants, we found that the growth-inhibitory effects in vitro were recovered by exogenous addition of 0.1 mM asparagine (Figure 3, D–G and Supplementary Figure S5A). Therefore, we examined whether the physiological concentration of asparagine (i.e., 0.05 mM) could recover the growth-inhibitory effects and found that 0.05 mM asparagine was not sufficient to recover the growth inhibition in vitro (Supplementary Figure S5C). In addition, the tumor cells within solid tumors in vivo are under a state of poorer nutrient, especially in the regions distant from tumor blood vessels. L-Asp treatment could diminish serum asparagine levels of mice and humans down to 0.004 and 0 mM, respectively [22], [45]. ASNS induction by L-Asp treatment was reported to be the resistance mechanism of L-Asp in the treatment of ALL [46]. Currently, ASNS inhibition has only been tested in L-Asp–refractory cases of ALL. ASNS inhibition induced cell death of glioblastoma cell lines in culture only when exogenous asparagine was absent [20]. Such inhibitors may well exhibit a therapeutic effect in other cancers when used together with L-Asp. A potent ASNS inhibitor available for use in vivo has not been developed yet [20], [29], [30] and is desirable clinically. In the present study, we found that L-Asp alone was not effective to KRAS-mutant xenografts (Figure 5E) and that ASNS protein expression was upregulated upon treatment with in the L-Asp alone (Supplementary Figure S6, A and B). We also demonstrated that rapamycin significantly suppressed ASNS expression in vitro and in vivo (Figure 2A and Supplementary Figure S6, A and B) and that the combination of L-Asp plus rapamycin markedly suppressed tumor growth in vivo (Figure 5E). Taken together, these results suggest that the combination of L-Asp plus rapamycin might be effective even in CRC with elevated ASNS expression. L-Asp and a rapamycin derivative, temsirolimus, were approved by FDA for the treatment of ALL and renal cell carcinoma, respectively [27], [28], [47]. It is notable that the safety of L-Asp and temsirolimus has already been assessed in these targeted diseases. However, native L-Asp derived from Escherichia coli cannot be acceptable in adult solid cancer patients due to its side effect [41]. A new L-Asp encapsulated within erythrocytes appears to improve safety compared with the native L-Asp [48], and a clinical trial is ongoing to evaluate the new L-Asp in PDAC metastatic setting [32]. Furthermore, chemical targeting of asparagine availability can also have a therapeutic value in several types of solid tumors including CRC.
The following are the supplementary data related to this article.
Primer Sequence for qRT-PCT
Authors' Contributors
Conception and design: K. Kawada.
Development of methodology: K. Toda, M. Iwamoto.
Acquisition of data: K. Toda, M. Iwamoto, S. Inamoto.
Analysis and interpretation of data: K. Kawada, K. Toda, S. Inamoto.
Writing, review, and/or revision of the manuscript: K. Kawada, K. Toda, M. Iwamoto, S. Inamoto, T. Sasazuki, S. Shirasawa, S. Hasegawa.
Administrative, technical, or material support: T. Sasazuki, S. Shirasawa, S. Hasegawa.
Study supervision: K. Kawada, Y. Sakai.
Footnotes
Grant supports: This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from the Laboratory for Malignant Control Research, Medical Innovation Center, Kyoto University Graduate School of Medicine (to K. Kawada).
Potential conflict of interest: No potential conflicts of interest were disclosed.
References
- 1.Jonker D.J., O'Callaghan C.J., Karapetis C.S., Zalcberg J.R., Tu D., Au H.J., Berry S.R., Krahn M., Price T., Simes R.J. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 2.Karapetis C.S., Khambata-Ford S., Jonker D.J., O'Callaghan C.J., Tu D., Tebbutt N.C., Simes R.J., Chalchal H., Shapiro J.D., Robitaille S. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 3.Ye L.C., Liu T.S., Ren L., Wei Y., Zhu D.X., Zai S.Y., Ye Q.H., Yu Y., Xu B., Qin X.Y. Randomized controlled trial of cetuximab plus chemotherapy for patients with KRAS wild-type unresectable colorectal liver-limited metastases. J Clin Oncol. 2013;31:1931–1938. doi: 10.1200/JCO.2012.44.8308. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L., Kepp O., Vander Heiden M.G., Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 8.Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J.K., Markowitz S., Zhou S. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawada K., Nakamoto Y., Kawada M., Hida K., Matsumoto T., Murakami T., Hasegawa S., Togashi K., Sakai Y. Relationship between 18F-fluorodeoxyglucose accumulation and KRAS/BRAF mutations in colorectal cancer. Clin Cancer Res. 2012;18:1696–1703. doi: 10.1158/1078-0432.CCR-11-1909. [DOI] [PubMed] [Google Scholar]
- 11.Iwamoto M., Kawada K., Nakamoto Y., Itatani Y., Inamoto S., Toda K., Kimura H., Sasazuki T., Shirasawa S., Okuyama H. Regulation of 18F-FDG accumulation in colorectal cancer cells with mutated KRAS. J Nucl Med. 2014;55:2038–2044. doi: 10.2967/jnumed.114.142927. [DOI] [PubMed] [Google Scholar]
- 12.Kawada K., Toda K., Nakamoto Y., Iwamoto M., Hatano E., Chen F., Hasegawa S., Togashi K., Date H., Uemoto S. Relationship between 18F-FDG PET/CT scans and KRAS mutations in metastatic colorectal cancer. J Nucl Med. 2015;56:1322–1327. doi: 10.2967/jnumed.115.160614. [DOI] [PubMed] [Google Scholar]
- 13.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBerardinis R.J., Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacevic Z., Morris H.P. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 1972;32:326–333. [PubMed] [Google Scholar]
- 16.Le A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C.J., Slusher B.S., Zhang H. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmerman L.A., Holton T., Yuneva M., Louie R.J., Padro M., Daemen A., Hu M., Chan D.A., Ethier S.P., van 't Veer L.J. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J.B., Erickson J.W., Fuji R., Ramachandran S., Gao P., Dinavahi R., Wilson K.F., Ambrosio A.L., Dias S.M., Dang C.V. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Fan J., Venneti S., Cross J.R., Takagi T., Bhinder B., Djaballah H., Kanai M., Cheng E.H., Judkins A.R. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirasawa S., Furuse M., Yokoyama N., Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 22.Hettmer S., Schinzel A.C., Tchessalova D., Schneider M., Parker C.L., Bronson R., Richards N.G., Hahn W., Wagers A.J. Functional genomic screening reveals asparagine dependence as a metabolic vulnerability in sarcoma. Elife. 2015;4 doi: 10.7554/eLife.09436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pencreach E., Guerin E., Nicolet C., Lelong-Rebel I., Voegeli A.C., Oudet P., Larsen A.K., Gaub M.P., Guenot D. Marked activity of irinotecan and rapamycin combination toward colon cancer cells in vivo and in vitro is mediated through cooperative modulation of the mammalian target of rapamycin/hypoxia-inducible factor-1alpha axis. Clin Cancer Res. 2009;15:1297–1307. doi: 10.1158/1078-0432.CCR-08-0889. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B., Dong L.W., Tan Y.X., Zhang J., Pan Y.F., Yang C., Li M.H., Ding Z.W., Liu L.J., Jiang T.Y. Asparagine synthetase is an independent predictor of surgical survival and a potential therapeutic target in hepatocellular carcinoma. Br J Cancer. 2013;109:14–23. doi: 10.1038/bjc.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz S., Wang J., Myeroff L., Parsons R., Sun L., Lutterbaugh J., Fan R.S., Zborowska E., Kinzler K.W., Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 26.Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 27.Chan W.K., Lorenzi P.L., Anishkin A., Purwaha P., Rogers D.M., Sukharev S., Rempe S.B., Weinstein J.N. The glutaminase activity of L-asparaginase is not required for anticancer activity against ASNS-negative cells. Blood. 2014;123:3596–3606. doi: 10.1182/blood-2013-10-535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narta U.K., Kanwar S.S., Azmi W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Crit Rev Oncol Hematol. 2007;61:208–221. doi: 10.1016/j.critrevonc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez J.A., Pan Y.X., Koroniak L., Hiratake J., Kilberg M.S., Richards N.G. An inhibitor of human asparagine synthetase suppresses proliferation of an L-asparaginase-resistant leukemia cell line. Chem Biol. 2006;13:1339–1347. doi: 10.1016/j.chembiol.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeuchi H., Ahn Y.M., Otokawa T., Watanabe B., Hegazy L., Hiratake J., Richards N.G. A sulfoximine-based inhibitor of human asparagine synthetase kills L-asparaginase–resistant leukemia cells. Bioorg Med Chem. 2012;20:5915–5927. doi: 10.1016/j.bmc.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 31.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen R., Neuzillet C., Tijeras-Raballand A., Faivre S., de Gramont A., Raymond E. Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget. 2015;6:16832–16847. doi: 10.18632/oncotarget.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J., Mullarky E., Lu C., Bosch K.N., Kavalier A., Rivera K., Roper J., Chio I.I., Giannopoulou E.G., Rago C. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;325:1555–1559. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scian M.J., Stagliano K.E., Deb D., Ellis M.A., Carchman E.H., Das A., Valerie K., Deb S.P., Deb S. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- 36.Ye J., Kumanova M., Hart L.S., Sloane K., Zhang H., De Panis D.N., Bobrovnikova-Marjon E., Diehl J.A., Ron D., Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasubramanian M.N., Butterworth E.A., Kilberg M.S. Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab. 2013;304:E789–E799. doi: 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards N.G., Kilberg M.S. Asparagine synthetase chemotherapy. Annu Rev Biochem. 2006;75:629–654. doi: 10.1146/annurev.biochem.75.103004.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui H., Darmanin S., Natsuisaka M., Kondo T., Asaka M., Shindoh M., Higashino F., Hamuro J., Okada F., Kobayashi M. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res. 2007;67:3345–3355. doi: 10.1158/0008-5472.CAN-06-2519. [DOI] [PubMed] [Google Scholar]
- 40.Lorenzi P.L., Llamas J., Gunsior M., Ozbun L., Reinhold W.C., Varma S., Ji H., Kim H., Hutchinson A.A., Kohn E.C. Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther. 2008;7:3123–3128. doi: 10.1158/1535-7163.MCT-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dufour E., Gay F., Aguera K., Scoazec J.Y., Horand F., Lorenzi P.L., Godfrin Y. Pancreatic tumor sensitivity to plasma L-asparagine starvation. Pancreas. 2012;41:940–948. doi: 10.1097/MPA.0b013e318247d903. [DOI] [PubMed] [Google Scholar]
- 42.Sircar K., Huang H., Hu L., Cogdell D., Dhillon J., Tzelepi V., Efstathiou E., Koumakpayi I.H., Saad F., Luo D. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. Am J Pathol. 2012;180:895–903. doi: 10.1016/j.ajpath.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., Zeller K.I., De Marzo A.M., Van Eyk J.E., Mendell J.T. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krall A.S., Xu S., Graeber T.G., Braas D., Christofk H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun. 2016;7:11457. doi: 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minowa K., Suzuki M., Fujimura J., Saito M., Koh K., Kikuchi A., Hamada R., Shimizu T. L-asparaginase-induced pancreatic injury is associated with an imbalance in plasma amino acid levels. Drugs R D. 2012;12:49–55. doi: 10.2165/11632990-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aslanian A.M., Fletcher B.S., Kilberg M.S. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J. 2001;357:321–328. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiarini F., Evangelisti C., McCubrey J.A., Martelli A.M. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36:124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Domenech C., Thomas X., Chabaud S., Baruchel A., Gueyffier F., Mazingue F., Auvrignon A., Corm S., Dombret H., Chevallier P. L-asparaginase loaded red blood cells in refractory or relapsing acute lymphoblastic leukaemia in children and adults: results of the GRASPALL 2005-01 randomized trial. Br J Haematol. 2011;153:58–65. doi: 10.1111/j.1365-2141.2011.08588.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer Sequence for qRT-PCT