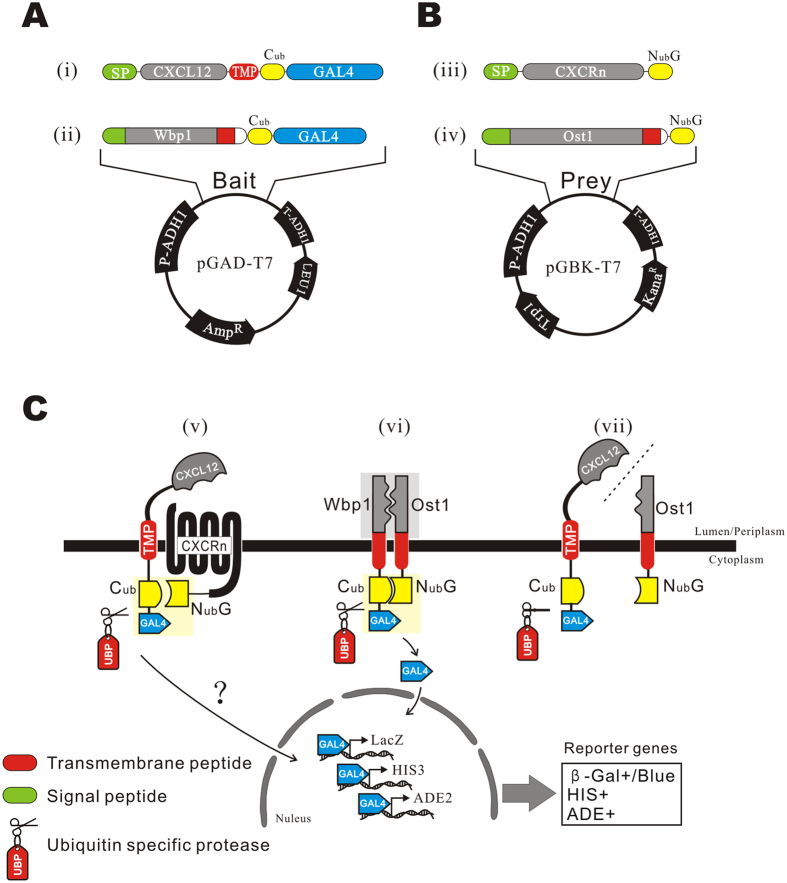
Figure 1. Design of bait and prey plasmids and the principles of detecting the interaction.
(A) The structures of the bait plasmids. (i) CXCL12 was fused with a signal peptide (SP) at the N-terminus and with a transmembrane peptide (TMP) at C-terminus, followed by Cub and GAL4 in tandem. (ii) WBP1 was fused with Cub and GAL4 as a control. Bait genes were cloned into the plasmid pGAD-T7. (B) Structures of prey plasmids. (iii) The prey fusion was composed of a SP, CXCR and NubG in tandem. (iv) OST1 was fused with NubG as a control prey. Prey genes were cloned into the plasmid pGBK-T7. (C) Principles of detecting the interaction. (v) The bait protein is expressed as a transmembrane protein, in which the CXCL12 domain is directed into the lumen (and the periplasm, if mature forms are located on plasmalemma) and linked with the Cub-GAL4 cassette in the cytoplasm by the TMP. CXCRs-NubG is co-expressed on the plasma membrane. The interaction of CXCL12 and CXCRs unites Cub and NubG and leads to the reconstitution of the split ubiquitin protein, which is recognized and cleaved by UBP to release GAL4. The liberated GAL4 protein is transported into the nucleus by the SV40 nuclear localization signal at its N-terminus, and transcription of reporter genes lacZ, HIS3 and ADE2 results in blue clones on a SD/His−Ade− plate in the presence of α-X-Gal. (vi) Coexpression of WBP1-Cub-GAL4 and OST1-NubG was used as the positive interacting proteins control. The two genes are known interacting partners of membrane proteins of yeast. (vii) Coexpression of SP-CXCL12-TMP-Cub-GAL4 and OST1-NubG, two unrelated genes, was used as a negative control.