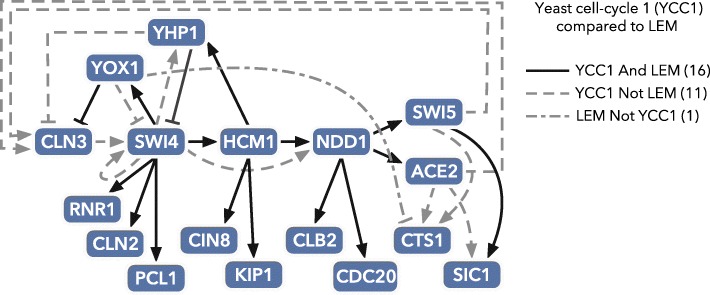
Fig. 4.
Using data from the budding yeast Saccharomyces cerevisiae and biological priors, LEM identifies a core cell-cycle transcription factor network. The topology of the yeast cell-cycle network was manually curated using literature evidence (see Additional file 1: Section 11). We ran LEM with biological prior information (see Additional file 1: Table 2), and we considered an edge to be output by LEM if had a posterior probability of at least 0.4. Edges present in both the cell-cycle network and the LEM output appear in bold. Edges that appear in the cell-cycle network but not in the LEM output are dashed. The edge that appears in the LEM output but not in the cell-cycle network is dash-dotted. LEM Local Edge Machine, YCC yeast cell cycle