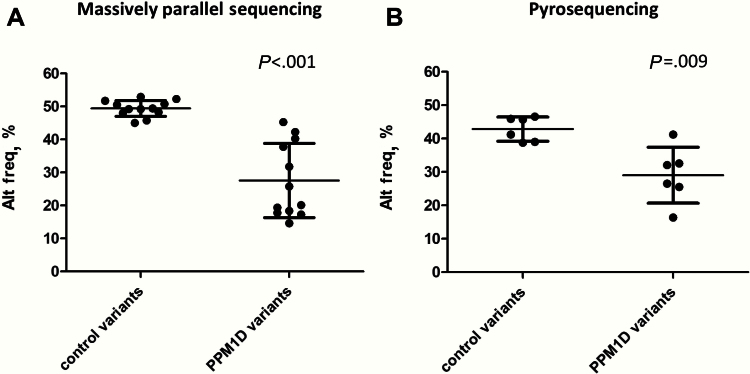
Figure 1.
Alternate allele frequency of PPM1D and other variants for the 13 samples with PPM1D truncating variants. A) Alternate allele frequency data from massively parallel sequencing. For the PPM1D variants, the frequency for each sample was graphed. For the other variants in 18 genes, the average of all identified variants in each individual, after homozygous variants were removed (alt freq >85%), was graphed. B) Alternate allele frequency data from pyro-sequencing for six cases with massively parallel sequencing alternate allele frequencies higher than 25%. For the PPM1D variants, the data from three to six replicates for each sample were averaged. Each of the six cases was heterozygous for either the single nucleotide polymorphism (SNP) BARD1 c.1134C>G or BRIP1 c.2637A>G. The data from three to six replicates of the control single-nucleotide polymorphism for each sample was averaged. Paired t tests were used to obtain two tailed P values.