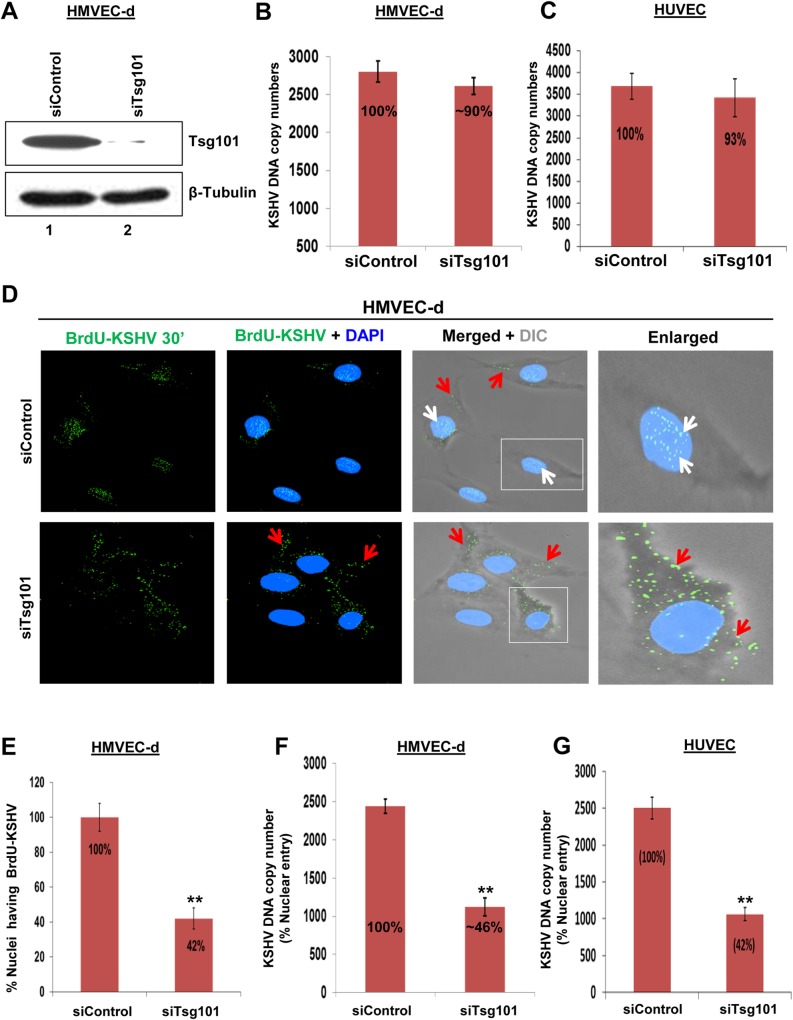
Fig 1. Effect of Tsg101 knockdown on KSHV entry in endothelial cells.
(A) HMVEC-d cells were transfected with siControl or siTsg101-RNA and examined after 48 h by Western blotting for Tsg101 and β-Tubulin. (B, C) siControl and siTsg101-RNA transfected HMVEC-d and HUVEC cells were infected with KSHV (30 DNA copies/cell) for 2 h at 37°C, washed and treated with 0.25% trypsin-EDTA for 5 min to remove the bound and non-entered virus. The cells were then treated with DNAse I for 10 min at 37°C according to the manufacturer's protocol to remove any contaminating DNA and assess only the viral DNA that was internalized. Total DNA was extracted and entry was determined by real-time DNA PCR for the KSHV ORF73 gene. The percentage of KSHV entry was calculated by considering the siControl value as 100%. Each reaction was done in triplicate and results presented are means ± SD of three independent experiments. (D) siControl and siTsg101-RNA transfected HMVEC-d cells were infected with BrdU (viral DNA) labeled KSHV (30 DNA copies/cell) for 30 min at 37°C, washed, treated with trypsin-EDTA for 5 min at 37°C, fixed and permeabilized using 0.2% Triton X-100 in PBS for 5 min, washed and followed by a denaturation step with 4N HCl for 10 min at RT to expose incorporated BrdU residues, washed, reacted with anti-BrdU antibodies followed by Alexa 488-anti-mouse antibodies and examined by immunofluorescence assay (IFA). Representative images are shown. White and red arrows indicate BrdU-KSHV genome associated with infected cell nuclei and cytoplasm, respectively. Magnification, 40x. (E) The percentage of nuclei positive for BrdU staining is presented graphically. A minimum of five independent fields, each with at least 10 cells was chosen. Results presented are means ± SD of three independent experiments. **p<0.01. (F, G) siControl and siTsg101-RNA transfected HMVEC-d and HUVEC cells were infected with KSHV (30 DNA copies/cell) for 2 h at 37°C, washed, treated with 0.25% trypsin-EDTA for 5 min at 37°C, washed, treated with DNAse I for 10 min at 37°C, washed, lysed on ice for 5 min with a mild lysis buffer, centrifuged at 500xg for 5 min to obtain the concentrated nuclear pellet. The cytoskeletal components loosely bound to the nuclei were removed by a second cycle of lysis and centrifugation steps [14]. The pure nuclear fraction was then used for DNA isolation and nuclear entry of the KSHV genome was determined by real-time DNA-PCR for the KSHV ORF73 gene. The percentage nuclear entry of genome was calculated considering the siControl value as 100%. Each reaction was done in triplicate. Results presented are means ± SD of three independent experiments. **p<0.01.