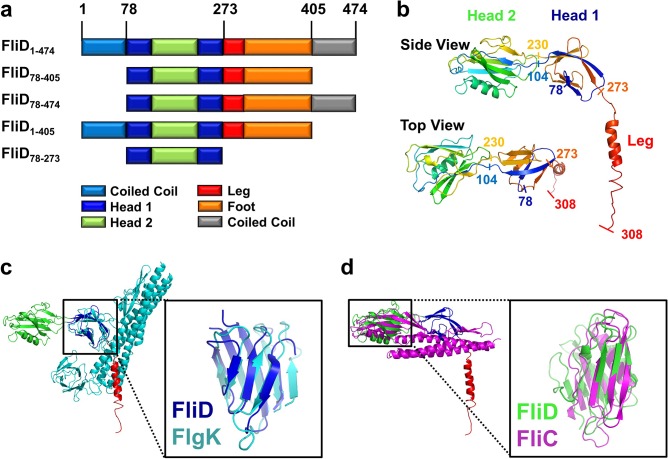
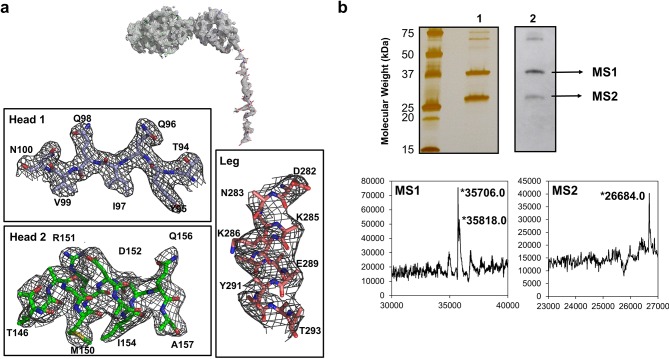
Figure 1. Crystal structure of Pseudomonas FliD reveals structural similarity to other flagellar proteins.
(a) Schematic representation of the FliD proteins used in these studies. Protein domain/region boundaries are labeled and are drawn approximately to scale. (b) Crystal structure of the Pseudomonas FliD78–405 monomer subunit with spectrum coloring from the N-terminus (blue) to the C-terminus (red). Head domain 1, head domain 2 and the leg region are indicated. (c) Superposition of the FliD78–405 crystal structure (domain coloring as in panel (a)) and Burkholderia FlgK/HAP1/hook filament capping protein (cyan). (d) Superposition of the FliD78–405 crystal structure (domain coloring as in panel (a)) and Pseudomonas flagellin/FliC (magenta).
DOI: http://dx.doi.org/10.7554/eLife.18857.003