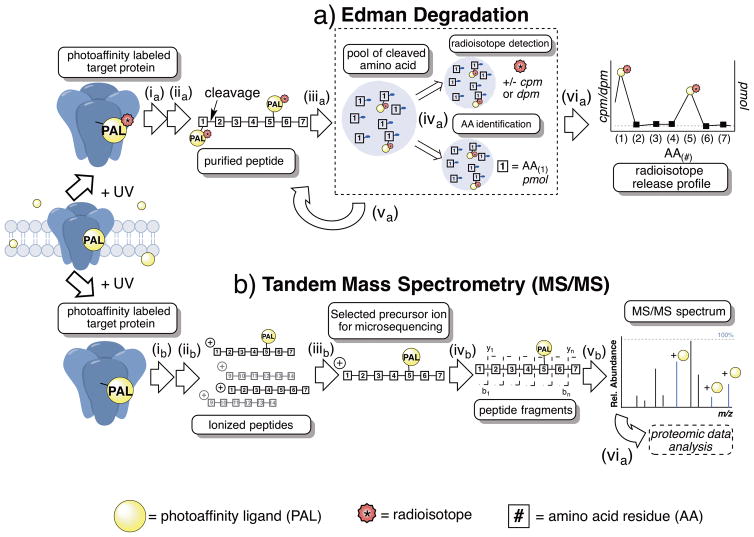
Figure 2.
Simplified schemes for the major methods used in micro-level anesthetic photoaffinity labeling; a) Edman degradation b) Tandem mass spectrometry (MS/MS). a) Edman degradation; ia) enriched target protein is photoaffinity labeled with radiolabeled (*) anesthetic photoaffinity ligand (PAL) and protein(s) are separated and digested into peptides; iia) peptides are isolated and rigorously purified; iiia) the first amino acid from the N-termini is cleaved from purified peptide via Edman reaction; iva) the cleaved amino acid is isolated, quantified and separated into two pools for (top) radioisotope detection by scintillation counting and (bottom) amino acid identification by chromatography; va) the cycle is repeated, gradually sequencing the entire purified peptide; via) the amino acid sequence is plotted against radioactivity (cpm, counts per minute, or dpm, disintegrations per minute) resulting in radioisotope release profile. b) Tandem mass spectrometry (MS/MS); ib) enriched target protein is photoaffinity labeled with anesthetic photoaffinity ligand (PAL) and protein(s) are separated and digested into peptides that are further separated by on- and/or offline chromatography methods; iib) peptides undergo ionization generally by electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI); iiib) precursor ions are separated by mass/charge (m/z) within MS1 (aka. MS1 precursor ion); ivb) MS1 precursor ion undergoes mass fragmentation often by collision-induced dissociation (CID) resulting in largely b or y fragment ions; vb) mass spectrometry software are used for data acquisition, database search analysis and representation.