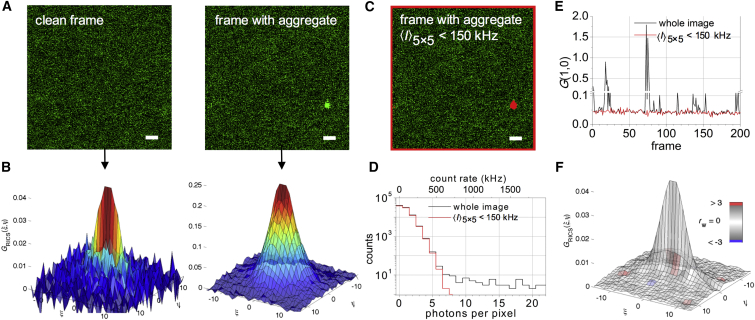
Figure 2.
Cleaning up image series before RICS analysis. (A) Two image frames from a 200-frame confocal image series of freely diffusing Venus fluorescent protein are shown. In the right image frame, a protein aggregate was in focus while the laser scanned over it. Scale bars represent 1 μm. (B) SACFs were calculated from the frames in (A). The aggregate dictates the shape of the second SACF. (C) The frame containing the aggregate, with the pixels around which a 5 × 5 pixel2 area exhibited an average intensity exceeding 150 kHz depicted in red. To avoid edge effects during thresholding, the borders of the image were also omitted from the ROI. The scale bar represents 1 μm. (D) Intensity distribution histogram is provided of (black) all pixels in the frame containing the aggregate and (red) all pixels included in the arbitrary ROI of the same frame. For comparison, both the absolute photon counts and the count rate per pixel are displayed. (E) The amplitudes of the individual SACFs were calculated (black) using all image pixels with Eq. 2 or (red) using all pixels included in the arbitrary ROI with Eq. 6. Amplitudes were approximated by the correlation value at ξ = 1. For clarity, a break at 0.11–0.2 was introduced on the y-axis. (F) The averaged arbitrary-ROI SACF was fitted using Eq. 14 and color-coded using the weighted residuals goodness-of-fit parameter rw. Fig. S1 contains further details on the fitting, as well as a comparison with the raw-data average SACF fitting. To see this figure in color, go online.