Abstract
Anthocyanin accumulation is responsible for flower coloration in peach. Here, we report the identification and functional characterization of eight flavonoid-related R2R3-MYB transcription factors, designated PpMYB10.2, PpMYB9, PpMYBPA1, Peace, PpMYB17, PpMYB18, PpMYB19, and PpMYB20, respectively, in peach flower transcriptome. PpMYB10.2 and PpMYB9 are able to activate transcription of anthocyanin biosynthetic genes, whilst PpMYBPA1 and Peace have a strong activation on the promoters of proanthocyanin (PA) biosynthetic genes. PpMYB17-20 show a strong repressive effect on transcription of flavonoid pathway genes such as dihydroflavonol 4-reductase. These results indicate that anthocyanin accumulation in peach flower is coordinately regulated by a set of R2R3-MYB genes. In addition, PpMYB9 and PpMYB10.2 are closely related but separated into two groups, designated MYB9 and MYB10, respectively. PpMYB9 shows a strong activation on the PpUGT78A2 promoter, but with no effect on the promoter of PpUGT78B (commonly called PpUFGT in previous studies). In contrast, PpMYB10.2 is able to activate the PpUFGT promoter, but not for the PpUGT78A2 promoter. Unlike the MYB10 gene that is universally present in plants, the MYB9 gene is lost in most dicot species. Therefore, the PpMYB9 gene represents a novel group of anthocyanin-related MYB activators, which may have diverged in function from the MYB10 genes. Our study will aid in understanding the complex mechanism regulating floral pigmentation in peach and functional divergence of the R2R3-MYB gene family in plants.
Keywords: peach, anthocyanin, proanthocyanin, flower coloration, R2R3-MYB
Introduction
Flower coloration is one of the most important traits in ornamental plants. In general, pigmentation in flowers is due to accumulation of various classes of pigments, such as anthocyanins, chlorophylls, carotenoids, and betalains (Tanaka et al., 2008). Anthocyanins are a kind of widely distributed water-soluble pigments, which are ultimately synthesized from phenylalanine in the cytosolic surface of the endoplasmic reticulum and stored in the vacuole. Anthocyanins are glycosylated forms of anthocyanidins catalyzed by glycosyltransferase in species-specific manner, with the sugar moiety predominantly attached at the 3-position on the C-ring or the 5-position on the A-ring (Caputi et al., 2012). Although more than 550 anthocyanins have been identified in nature, there are only six major types of anthocyanidins: pelargonidin, cyanidin, peonidin, delphinidin, petunidin, and malvidin (Kong et al., 2003). The color change of anthocyanidins is mainly attributed to the number of hydroxyl groups on the B-ring, with more hydroxyl groups shifting the color to blue. Glycosylation of anthocyanidins has a slight reddening effect (Tanaka et al., 2008). Thus, understanding mechanism regulating anthocyanin biosynthesis is crucial for genetic manipulation of flower coloration in ornamental plants.
The anthocyanin biosynthetic pathway has been elucidated in a variety of plant species (Grotewold, 2006). This pathway involves a number of reactions, which are catalyzed by various enzymes such as chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′ hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT). Of these enzymes, F3′5′H is absent in some plants such as Arabidopsis (Forkmann, 1991), apple (Han et al., 2010), and peach (Cheng et al., 2014), and each is encoded by gene families. For example, two UFGT78 genes, PpUGT78A2 and PpUGT78B, are identified in the peach genome, and both of them can catalyze the glycosyl transfer on the C3 position of the anthocyanidin aglycone in flower (Cheng et al., 2014).
Besides anthocyanin pathway structural genes, various regulatory genes, such as MYB, basic helix-loop-helix (bHLH), and WD40 transcription factors (TFs), are also involved in anthocyanin biosynthesis. Although these three types of TFs form a complex to coordinately regulate the transcription of anthocyanin pathway structural genes, MYB TFs such as Arabidopsis AtPAP1/AtMYB75 (Borevitz et al., 2000) or its homologs are widely reported to play a determinant role in anthocyanin coloration in various dicotyledonous plants, such as Antirrhinum majus (Schwinn et al., 2006), cauliflower (Chiu et al., 2010), Petunia (Albert et al., 2011), kiwifruit (Fraser et al., 2013), Asiatic hybrid lily (Yamagishi et al., 2014), monkeyflowers (Yuan et al., 2014), and Phalaenopsis spp. (Hsu et al., 2015).
Peach belongs to the genus Pruns in the Rosaceae family. It is a diploid, with a basic chromosome number of x = 8. Peach has a small genome of approximately 230 Mb per haploid and its draft genome sequence was released in 2010 (Verde et al., 2013). More recently, several anthocyanin-related MYB genes have been identified to control anthocyanin accumulation in peach. For example, three R2R3-MYB genes, PpMYB10.1, PpMYB10.2, and PpMYB10.3, are involved in anthocyanin accumulation in fruit (Ravaglia et al., 2013; Rahim et al., 2014; Zhou et al., 2015b), while the PpMYB10.4 gene is responsible for red coloration of leaf (Zhou et al., 2014). A TT2-like R2R3-MYB gene, designated Peace, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers although it is unclear whether the Peace gene is able to activate anthocyanin and/or proanthocyanin pathway structural genes (Uematsu et al., 2014). However, our study shows that a small indel mutation in an anthocyanin transporter, which is not the result of a transposon insertion or excision, causes variegated colouration of flowers of ornamental peach, ‘Hongbaihuatao’ (Cheng et al., 2015). These results suggest a complex mechanism regulating anthocyanin accumulation in peach flower.
We previously conducted RNA-Seq for different tissues of peach, including leaf, flower, and fruit (Wang et al., 2013). Here, we analyzed the RNA-Seq transcriptome of peach flower, and a total of eight flavonoid-related R2R3-MYB TFs, designated PpMYB10.2, PpMYB9, PpMYBPA1, Peace, PpMYB17, PpMYB18, PpMYB19, and PpMYB20, were identified. Among these MYB TFs, PpMYB10.2, and PpMYB9 are anthocyanin activators, while PpMYBPA1 and Peace are involved in the regulation of proanthocyanin (PA) biosynthesis. The rest four MYBs are flavonoid repressors. Interestingly, the PpMYB9 gene showed the highest level of expression in flower and it represents a novel R2R3-MYB family regulating specific UFGT gene in peach. Our study reveals a set of R2R3-MYBs involved in the regulation of anthocyanin accumulation in peach flower, which will be helpful for manipulating anthocyanin coloration in peach programs in the future.
Materials and Methods
Plant Materials
Peach accessions used for transcriptome analysis in this study contained one cultivar Dahongpao and two ornamental cultivars, Hongbaihua and Hongyetao. These three cultivars are maintained at Wuhan Botanical Garden of the Chinese Academy of Sciences (Beijing, China). Blood-fleshed fruits of ‘Dahongpao,’ red flowers of ‘Hongbaihua,’ and red leaves of ‘Hongyetao’ were collected at 85 days after full-bloom (DAFB), flower bud stage, and juvenile stage in spring season, respectively. For cv. Mantianhong, flower samples were collected at flower bud, balloon and full-balloon stages, respectively. All the samples were immediately frozen in liquid nitrogen and then stored at -80°C until use.
Identification of R2R3-MYB Genes in Peach Transcriptome
Details about cDNA library preparation, Illumina sequencing, and the mapping, assembly and GO (Gene Ontology) annotation of RNA-Seq data were present in our previous study (Wang et al., 2013). To validate the assembled unigenes, all unigenes were BLASTed against GenBank’s non-redundant protein (NR) and Swiss-Prot protein databases using BLASTx, and against GenBank’s non-redundant nucleotide (NT) database using BlastN, with an E-value cutoff of 1e-5. The FPKM values of R2R3-MYB genes showing high-level expression (FPKM ≥ 3) in one or more tissues tested were selected for further analysis. The FPKM value was used to create the heatmap using MeV (Multiple Experiment Viewer) 4.81 software (Howe et al., 2011).
Construction of Expression Vectors
A pair of primers, MYB9F and MYB9R, was designed to amplify the full coding region of PpMYB9 using KOD-Plus neo kit (Toyobo, Osaka, Japan). PCR products were purified, digested using restriction enzymes EcoRI and HindIII (New England Biolabs, Beverly, MA, USA), and inserted into the multiple cloning site (MCS) of pSAK277 binary vector. Similarly, the coding sequences of PpMYB10.2, PpMYBPA1, Peace, PpMYB17, PpMYB18, PpMYB19, and PpMYB20 were cloned and inserted individually into the MCS of pSAK277, while the PpbHLH3 gene was cloned and inserted into binary vector pHEX2 according to Zhou et al. (2015a). proUGT78A2::UGT78A2 was constructed in two steps: UGT78A2 was inserted into the MCS of pSAK277 vector, followed with replacement of CaMV 35S promoter by the promoter of PpUGT78A2 (1.8 Kb). The primers used for quantitative real-time PCR (qRT-PCR) are listed in Supplementary Table S1.
Dual Luciferase Reporter Assay
A transient dual luciferase reporter assay was conducted in young leaf of tobacco (Nicotiana benthamiana) according to previously reported protocol (Hellens et al., 2005; Espley et al., 2007). Promoter regions upstream from the start codon of six peach genes, DFR (1.6 Kb), LDOX (1.9 Kb), LAR1 (1.4 Kb), ANR (1.9 Kb), UFGT (2.5 Kb), and UGT78A2 (1.8 Kb), were isolated and inserted individually into the MCS of vector pGreenII 0800-LUC. All the constructs were individually transformed into A. tumefaciens strain GV3101, and incubated at 28°C for 2 days. The confluent bacteria were resuspended in 10 ml infiltration buffer containing 10 mM MgCl2, 200 μM acetosyringone and 10 mM 2-(N-morpholine)-ethanesulfonic acid (pH = 5.7). Before infiltration, the bacteria were incubated at room temperature without shaking for 2 h. Transient transformation was conducted by mixing Agrobacterium GV3101 culture transformed with the reporter cassette and with the cassettes containing PpMYBs and PpbHLH3 driven by the CaMV 35S promoter with a ratio of 2:9:9. All tests were repeated at least four biological replicates. Firefly luciferase (Luc) and renilla luciferase (Ren) activity were measured 3 days after infiltration using Dual-Glo® Luciferase Assay System (Promega, Madison, WI, USA) on an Infinite M200 luminometer (Tecan, Mannerdorf, Switzerland). Promoter activation activity was shown as the ratio of Luc activity to Renilla activity (Luc/Ren).
Induction of Anthocyanins by Transient Infiltration in Nicotiana tabacum Leaf
Young tobacco seedlings (2–4 week old) were used for transient color assay. Agrobacteria cultivation and infiltration preparation were carried out following the same protocol as described above for transient dual luciferase assay. Equivalent doses of PpMYB9, PpbHLH3, and proUGT78A2::UGT78A2 were mixed and infiltrated into the abaxial side of young leaves. The seedlings were placed in darkness overnight, and then moved to glasshouse under a long day photoperiod (16 h light/8 h dark). Photographs were taken at 2 weeks after infiltration.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using Total RNA Rapid Extraction Kit (Zomanbio, Beijing, China). Removal of genomic DNA contamination and first strand cDNA synthesis were conducted using PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Quantitative RT-PCR was carried out in a total reaction volume of 20 μL containing 100 ng of template cDNA, 0.4 μM of each primer, 1 × ROX reference dye, and 10 μL of 2 × SYBR premix Ex Taq II (TaKaRa). The qRT-PCR program was as follows: one cycle of 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 34 s at 60°C. A translation elongation factor gene PpTEF2 was selected as an internal control according to the previous report (Tong et al., 2009). All analyses were repeated three times using three biological replicates. The primers used for qRT-PCR are listed in Supplementary Table S2.
Results
Identification of Flavonoid-Related R2R3-MYB Transcription Factors in Peach Flower Transcriptome
In our previous study (Wang et al., 2013), we reported an RNA-Seq-based transcriptome database of peach. RNA-Seq data analysis revealed 148 transcripts encoding R2R3-MYB TFs. The expression level of these R2R3-MYB TFs was estimated in different tissues, including red flower, red leaf, and blood fruit. As a result, 49 R2R3-MYB TFs were identified to have relatively high expression levels (FPKM ≥ 3) in at least one of the three tissues tested. Subsequently, a heatmap representing their expression profiles was constructed based on the values of FPKM (fragments per kilobase of transcript per million mapped reads; Figure 1). Overall, the 49 R2R3-MYBs showed extensive variation in expression among the tested tissues. For example, PpMYB10.1 (ppa026640m) was only expressed in blood mesocarp, while the transcript of PpMYB10.4 (ppa018744m) was only observed in red leaf. Of the 49 R2R3-MYBs, eight, PpMYB10.2 (ppa016711m), PpMYB9 (ppa010069m), PpMYBPA1 (ppa009439m), Peace (ppa023768m), PpMYB17 (ppa010277m), PpMYB18 (ppa010846m), PpMYB19 (ppa010716m), and PpMYB20 (ppa008906m), showed relatively high-level expression in flower. These eight R2R3-MYBs were selected and subjected to later analysis to elucidate mechanisms regulating anthocyanin biosynthesis in peach flower.
FIGURE 1.
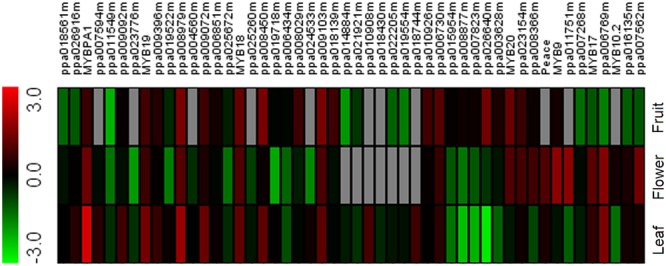
Heatmap of R2R3-MYBs in peach transcriptomes of different red-colored tissues. Green and red boxes indicate low- and high-expression levels, respectively. Red-colored leaf, flower, and fruit samples were collected from cv. Hongyetao, Hongbaihua, and Dahongpao, respectively.
Phylogenetic analysis showed that all the R2R3-MYBs except PpMYB9 were clustered with known function MYBs of flavonoid biosynthesis (Figure 2). For example, PpMYB10.2 was closely related to anthocyanin MYB activators such as MdMYB10 and PpMYB10.1, while PpMYBPA1 and Peace were similar to PA-related MYBs, VvMYBPA1 and VvMYBAP2, respectively. PpMYB17-20 was closely related to flavonoid MYB repressors such as FaMYB1 and VvMYBC2-L1. In contrast, PpMYB9 along with other unknown function MYBs formed a new group, which was phylogenetically related to anthocyanin MYB activators. Moreover, we further checked the MYB9 gene in sequenced genomes of 24 plant species, and found that it is present in only seven species, including soybean, Lotus japonicus, Medicago truncatula, apple, Eucalyptus grandis, cotton, and papaya (Table 1).
FIGURE 2.
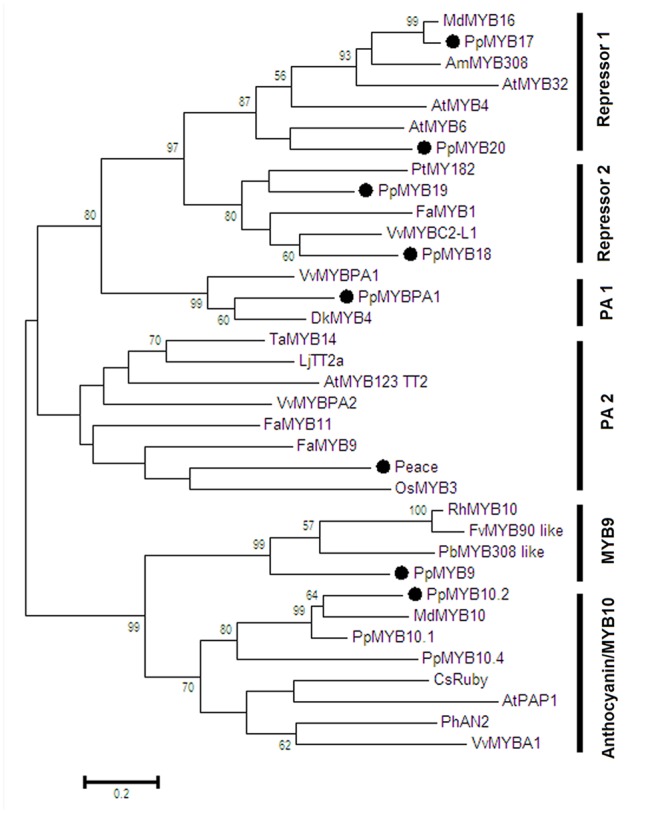
Phylogenetic tree derived from amino acid sequences of R2R3-MYBs in peach and other dicot species. The peach R2R3-MYBs are highlighted with a solid black circle. Full amino acid sequences of R2R3-MYBs were aligned using Muscle software, and phylogenetic tree was constructed with MEGA software (version 6.0) using maximum likelihood method. Numbers near branches indicate bootstrap values that were calculated from 1000 replicate analyses. The scale bar represents 0.2 substitutions per site. The accession numbers are as follows: Arabidopsis thaliana AtMYB4 (AT4G38620), AtMYB6 (AT4G09460), AtPAP1 (AT1G56650), AtMYB123 (AT5G35550), AtMYB32 (AT4G34990); Vitis vinifera VvMYBA1 (AB097923), VvMYBPA2 (EU919682), VvMYBPA1 (AM259485), VvMYBC2-L1 (JX050227); Prunus persica PpMYB10.2 (EU155160), PpMYBPA1 (CV047374), Peace (AB897865), PpMYB9 (KT159232), PpMYB17 (KT159233), PpMYB18 (KT159234), PpMYB19 (KT159235), PpMYB20 (KT159236), PpMYB10.4 (KF999985), PpMYB10.1 (XM_007216468); Malus domestica MdMYB10 (DQ267897), MdMYB16 (HM122617); Fragaria ananassa FaMYB9 (JQ989281), FaMYB11 (JQ989282), FaMYB1 (AF401220); Oryza sativa OsMYB3 (D88619); Diospyros kaki DkMYB4 (AB503701); Trifolium arvense TaMYB14 (JN049641); Petunia x hybrid PhAN2 (ABO21074); Citrus sinensis CsRuby (NM_001288889); Lotus japonicus LjTT2a (AB300033); Antirrhinum majus AmMYB308 (P81393); Populus tremula x Populus tremuloides PtMYB182 (KP723392); Rosa hybrida RhMYB10(EU155164); Fragaria vesca FvMYB90 like(XM_004302074); Pyrus x bretschneideri PbMYB208 like (XM_009339740).
Table 1.
Copy numbers of MYB9 gene in the sequenced genomes of eudicots.
| Eudicots | Species | MYB9 | ||
|---|---|---|---|---|
| Rosids | Caryophyllales | Amaranthaceae | Beet | 0 |
| Vitales | Vitaceae | Grapevine | 0 | |
| Eudicots I | Salicaceae | Poplar | 0 | |
| Euphorbiaceae | Cassava | 0 | ||
| Ricinus | 0 | |||
| Fabaceae | Soybean | 2 | ||
| L. japonicus | 1 | |||
| M. truncatula | 1 | |||
| Rosaceae | Peach | 1 | ||
| Apple | 1 | |||
| Strawberry | 0 | |||
| Cucurbitaceae | Muskmelon | 0 | ||
| Watermelon | 0 | |||
| Eudicots II | Myrtaceae | E. grandis | 1 | |
| Rutaceae | Orange | 0 | ||
| Malvaceae | Cocoa | 0 | ||
| Cotton | 1 | |||
| Caricaceae | Papaya | 1 | ||
| Brassicaceae | T. parvula | 0 | ||
| B. rapa | 0 | |||
| C. rubella | 0 | |||
| A. lyrata | 0 | |||
| A. thaliana | 0 | |||
| Asterids | Solanaceae | Potato | 0 | |
| Tomato | 0 | |||
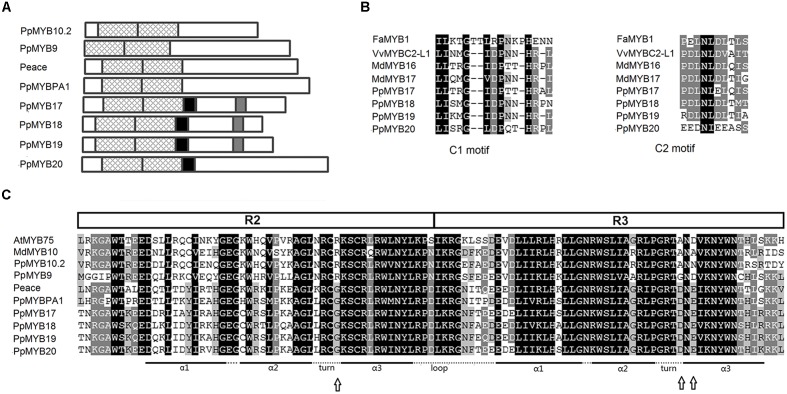
Sequence alignment revealed that all the eight MYBs contained conserved R2 (53 amino acids) and R3 (52 amino acid) domains (Figure 3A). However, only four repressor-like MYBs, PpMYB17 through PpMYB20, contained C1 and/or C2 motifs (Figures 3A,B). A residue at position 39 in the R2 domain and four residues at positions 90–93 in the R3 domain are known to be key amino acid elements that control the specificity of MYB regulators for either the anthocyanin or PA pathway (Heppel et al., 2013). Both PpMYBPA1 and Peace shared the same residue Gly at position 39 in the R2 domain and Asp-Asn-Glu-Ile/Val at positions 90–93 in the R3 domain (Figure 3C), which suggests that they are probably involved in PA pathway. Similarly, PpMYB10.2 and PpMYB9 contained the same residue Arg at position 39 in the R2 domain, but the residus at positions 90–93 of PpMYB9 (Gly-Asn-Asp-Val) were different from that of their counterpart in PpMYB10.2 (Ala-Asn-Asn-Val), as well as other anthocyanin MYB activators such as MdMYB10 and AtMYB75. In addition, the residue at position 99 was also different between PpMYB9 (Cys) and other anthocyanin MYB activators (Thr or Ser). These results suggest that the recognition site of PpMYB9 in the promoter of anthocyanin structural genes may differ from that of other anthocyanin MYB activators, leading to functional divergence between PpMYB9 and PpMYB10.2.
FIGURE 3.
Diagram and alignment of eight R2R3-MYB transcription factors (TFs). (A), structural diagram of the eight R2R3-MYBs highly expressed in peach flower. Cross lines indicate R2 and R3 SANT domains. The black and grey boxes stand for C1 and C2 motif, respectively. (B,C), amino acid sequence alignment of R2R3 domain and C1/C2 motif, respectively, of the MYBs in peach and other dicots. Arrows highlight key amino acid residues that mediate promoter targets specificity for the anthocyanin and proanthocyanin (PA) pathway (Heppel et al., 2013).
Functional Analysis of the Eight R2R3-MYBs Using Dual Luciferase Assay
To testify whether the eight R2R3-MYBs had direct activation on transcription of structural genes, the promoter regions of six flavonoid pathway genes in peach, DFR, LDOX, LAR1, ANR, UFGT, and UGT78A2, were isolated and inserted into dual luciferase assay system (Hellens et al., 2005). Four MYB genes (PpMYB10.2, PpMYB9, PpMYBPA1, and Peace) with the PpbHLH3 partner were all able to activate the promoters of early flavonoid pathway genes, with LUC/REN ratio ranging from 1.7 to 5.1 for the DFR promoter, and from 1.9 to 3.8 for the LDOX promoter (Figure 4). This suggests that these four MYB genes are probably involved in the regulation of anthocyanin or proanthocyanin biosynthesis in peach.
FIGURE 4.
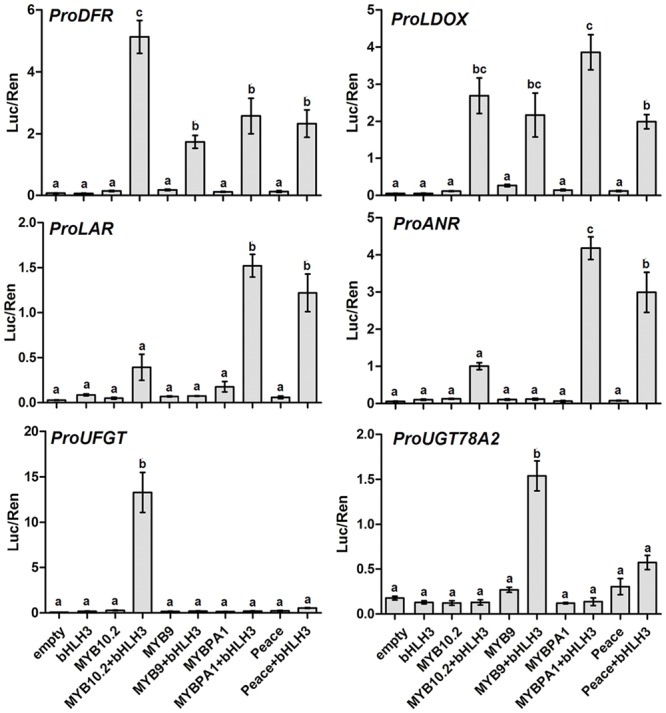
Estimation of effect of different R2R3-MYBs with PpbHLH3 partner on activation of the promoter of anthocyanin or PA pathway genes using transient dual luciferase assay. Promoter activity was shown as the ratio of firefly Luciferase (Luc) activity to Renilla luciferase (Ren) activity. Different lowercase letters indicate the significant differences among treatments by Tukey’s test at P < 0.01. Error bars show SE of four biological replicates.
Our previous study shows that cyanidin 3-glucoside is the main component of anthocyanins in peach flower, and two UFGT78 genes, PpUGT78B (also commonly called PpUFGT in previous studies) and PpUGT78A2, are involved in the glycosylation of cyanidin (Cheng et al., 2014). Here, dual luciferase assay further revealed that only PpMYB10.2 co-infiltrated with PpbHLH3 was able to activate the PpUFGT promoter, with a LUC/REN ratio of 13.3. However, either PpMYB10.2 or PpMYBPA1 co-infiltrated with PpbHLH3 showed a weak activation on the PpUFGTA2 promoter. The PpMYB9 gene with the PpbHLH3 partner showed the strongest activation on the PpUFGTA2 promoter, with a LUC/REN ratio of 1.5. In addition, Peace with the PpbHLH3 partner showed a twofold decrease in the PpUFGTA2 promoter activation when compared with PpMYB9, but its activity was approximately four or fivefold higher than that of PpMYBPA1.
In peach, two structural genes, PpLAR1 and PpANR, were found to be related to proanthocyanidin biosynthesis (Ravaglia et al., 2013). Here, our results indicated that both PpMYBPA1 and Peace with the PpbHLH3 partner displayed a strong activation on the PpLAR1 promoter, with the LUC/REN ratios of 1.5 and 1.2, respectively. Similarly, both PpMYBPA1 and Peace with the PpbHLH3 partner were also able to effectively activate the PpANR promoter, with the LUC/REN ratios of 4.2 and 3.0, respectively. PpMYB10.2 co-infiltrated with PpbHLH3 showed a slight activation on the promoters of PpLAR1 and PpANR, with the LUC/REN ratios of 0.4 and 1.0, respectively. However, no activation was observed for the infiltration of PpMYB9 alone or with the PpbHLH3 partner.
In addition, PpDFR promoter was selected to validate the function of the four flavonoid MYB repressors, PpMYB17-20. Co-infiltration of PpMYB10.2 and PpbHLH3 showed a strong activation on the DFR promoter (Figure 5). However, this activation was completely counteracted by addition of any of the four flavonoid MYB repressors.
FIGURE 5.
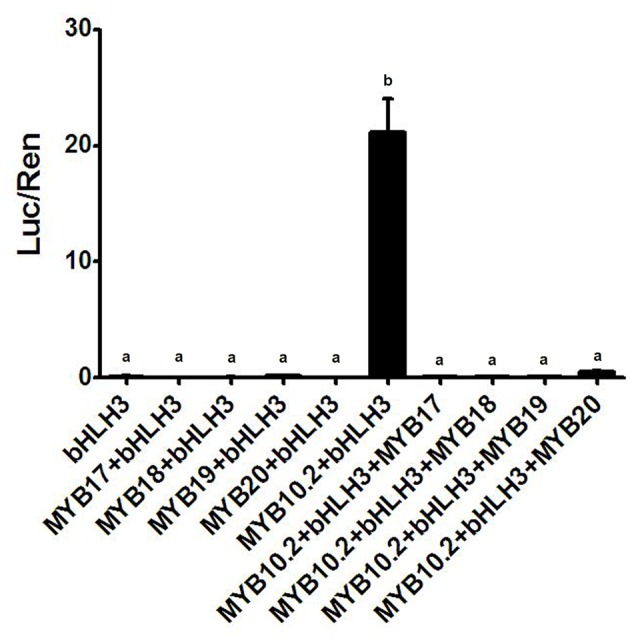
Validation of activation effect of four R2R3-MYBs on the PpDFR promoter using dual luciferase assay. Different lowercase letters indicate the significant differences among treatments by Tukey’s test at P < 0.001. Results represent the means of four biological replicates.
In general, the above results suggest that of the eight R2R3-MYBs expressed in peach flower, PpMYB10.2 and PpMYB9 are anthocyanin-related MYB activators, while PpMYBPA1 and Peace belong to proanthocyanin-related MYB regulators. However, the other four, PpMYB17-20, are flavonoid-related MYB repressors.
Transient Color Assays of the Peach MYB9 Activity
Since PpMYB9 represents a new MYB group as mentioned above, its function was further validated by transient color assay in N. tabacum leaf via Agrobacterium infiltration. Initially, PpMYB9 and PpbHLH3 were infiltrated into the abaxial side of tobacco leaf. However, no red pigmentation was observed in 2 weeks (Figure 6A). Considering the fact that PpMYB9 specifically activated the promoter of PpUGT78A2 instead of the common PpUFGT, a construct proPpUGT78A2::PpUGT78A2 was co-infiltrated with PpMYB9 and PpbHLH3. Red pigmentation was detected around the infiltration site in 2 weeks (Figure 6B). However, no pigmentation was observed for the infiltration of proPpUGT78A2::PpUGT78A2 alone (Figure 6C). These results indicated that heterologous expression of PpMYB9 and PpbHLH3 was able to induce anthocyanin accumulation via activating transcription of PpUGT78A2.
FIGURE 6.
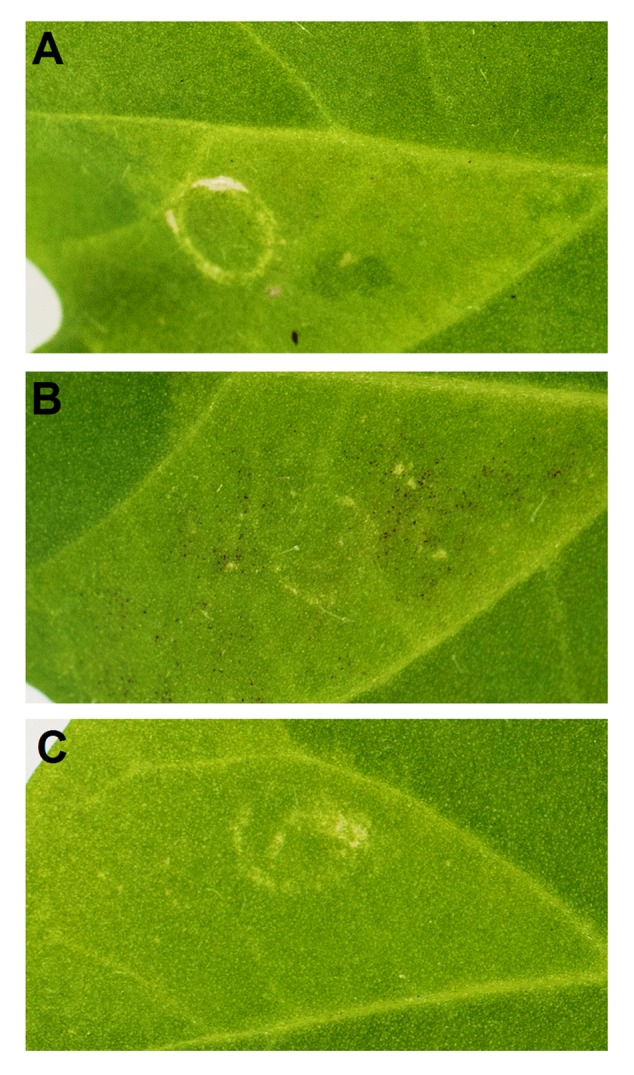
Transient color assay of the PpMYB9 activity in N. tabacum leaf. (A–C) indicate infiltration with PpMYB9+PpbHLH3, PpMYB9+PpbHLH3+proPpUGT78A2::PpUGT78A2, and proPpUGT78A2::PpUGT78A2, respectively. Photos were taken 2 weeks after infiltration.
Expression Profiles of the Eight R2R3-MYBs in Peach Flower
An ornamental peach variety ‘Mantianhong’ was selected to investigate the expression profile of the eight R2R3-MYBs in flower. Of the four MYB activators, PpMYB9 showed the highest level of expression throughout the developmental stages analyzed (Figure 7). The transcript level of PpMYB10.2 increased significantly during the process of flower development, with a high peak at full-bloom stage. Both PpMYBPA1 and Peace were expressed at bud stage, whereas, with almost undetectable transcript level at either balloon or full-bloom stages. Of the four MYB repressors, PpMYB17 and PpMYB20 were highly expressed at bud and bloom stages, but decreased significantly at full-bloom stage. The transcript level of PpMYB18 and PpMYB19 was relatively low and showed a significantly decreasing pattern during the process of flower development.
FIGURE 7.
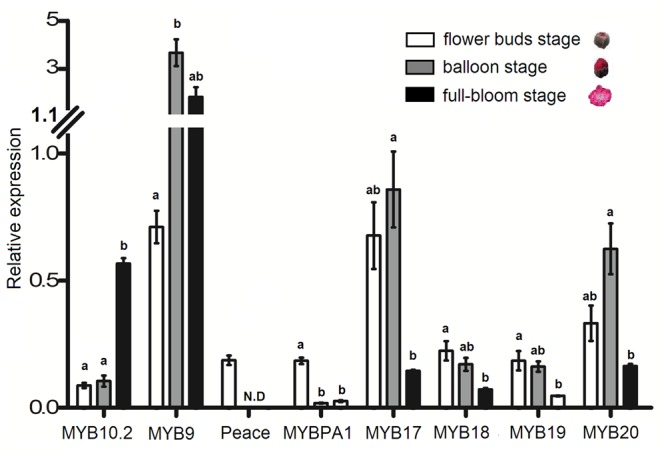
Expression profiles of the eight R2R3-MYBs in flowers of cv. Mantianhong at different developmental stages. Different lowercase letters indicate the significant differences among treatments by Tukey’s test at P < 0.01. Error bars show SE of the means of three biological replicates.
Discussion
Anthocyanin Accumulation in Peach Flower Is Regulated by a Set of R2R3-MYB Genes
In plants, anthocyanin biosynthesis is regulated by the MYB-bHLH-WD40 “MBW” complex. Of these three regulators, R2R3-MYBs play a crucial role in determining anthocyanin accumulation (Ramsay and Glover, 2005). It has been documented that multiple R2R3-MYB TFs are involved in the regulation of floral pigmentation in ornamental plants. For example, four R2R3-MYB TFs, AN2, AN4, PHZ, and DPL, control anthocyanin accumulation in different parts of petunia flower (Quattrocchio et al., 1998, 1999; Albert et al., 2011). Three R2R3-MYB TFs, Rosea1, Rosea2, and Venosa, are involved in the regulation of flower pigmentation in the genus Antirrhinum (Schwinn et al., 2006). Three R2R3-MYB TFs are responsible for distinct patterns of floral pigmentation in Orchidaceae (Hsu et al., 2015).
In this study, four R2R3-MYB activators, PpMYB9, PpMYB10.2, PpMYBPA1, and Peace, were found in peach flower transcriptome. Functional analysis demonstrated that PpMYB10.2 and PpMYB9 are related to anthocyanin biosynthesis, whilst PpMYBPA1 and Peace are mainly involved in regulation of PA biosynthesis. Both PpMYB10.2 and PpMYB9 are expressed in flower, with a high-level expression in late development stage. This suggests that PpMYB9 and PpMYB10.2 are crucial for floral coloration in peach. The high-levels of expression of these two MYB genes during might play an important role in maintenance of the flower coloration during late stages of flower development. Unlike the anthocyanin-related MYB activators, PpMYBPA1 and Peace show extremely low or undetectable levels of expression in flower at balloon and full-bloom stages, suggesting that they may play little role in peach floral coloration at the later developmental stages. A more recent study shows that Peace has high-level expression in flower bud of cv. Genpei and its expression change causes variegated pattern of floral coloration (Uematsu et al., 2014). Interestingly, Peace was shown to activate anthocyanin biosynthesis. However, our study indicates that Peace is phylogenetically related to PpMYBPA1, and both of them have a strong activation on the promoters of PA biosynthetic genes, PpLAR and PpANR. Overexpression of either LAR or ANR can result in loss of anthocyanin in flower (Han et al., 2012; Liao et al., 2015). Thus, the regulatory role of Peace in peach flower coloration needs further investigation.
It is worth noting that Peace belongs to PA regulator group, and its transient expression was shown to induce pigmentation in white flower of ‘Genpei’ (Uematsu et al., 2014). However, our study demonstrates that the Peace gene can efficiently activate PA-specific pathway genes such as LAR and ANR, whereas, with low activation of anthocyanin-specific pathway genes such as PpUGT78A2. This indicates that the Peace gene is unlikely to play an important role in anthocyanin pigmentation. More studies are still needed to address the mechanism by which the Peace gene regulates anthocyanin pigmentation in peach flower.
Besides anthocyanin-related MYB activators, four flavonoid-related MYB repressors, PpMYB17 through PpMYB20, are also found in peach flower transcriptome. All these MYB repressors have a strong repressive effect on flavonoid pathway genes such as DFR, and are expressed in flower during the whole process of development. These results indicate that PpMYB17-20 may play a negative role in the regulation of anthocyanin accumulation in peach flower.
Taken together, our study suggests that anthocyanin accumulation in peach flower is regulated by a set of R2R3-MYB genes, including four MYB activators and four MYB repressors. Coordinate regulation of multiple R2R3-MYB genes is likely responsible for flower color intensity variation in peach, ranging from white or pale to red, pink, and purple.
PpMYB9 Represents a New Group of R2R3-MYB Genes Regulating Anthocyanin Biosynthesis in Plants
In this study, phylogenetic analysis reveals that anthocyanin-related MYB activators are divided into two groups, MYB10 and MYB9 (Figure 2 and Supplementary Figure S1). The MYB10 group consists of the peach PpMYB10.2 and most previously reported R2R3-MYB activators controlling anthocyanin pigmentation in dicots, such as Arabidopsis AtPAP1 and AtPAP2 (Borevitz et al., 2000), apple MdMYB10 (Espley et al., 2007), citrus CsRuby (Butelli et al., 2012), Petunia × hybrid PhAN2 (Quattrocchio et al., 1998), AmVENOSA, Antirrhinum majus AmROSEA1 and AmROSEA2 (Schwinn et al., 2006), and grapevine VvMYBA1 and VvMYBA2 (Walker et al., 2007).
The MYB9 group that includes the peach PpMYB9 is different from the MYB10 group. Firstly, the MYB9 gene is lost in most dicot species (Table 1), whereas, the MYB10 gene is present in all eudicot species. This suggests that the MYB10 group represents a common anthocyanin-related MYB activator in plants, while the MYB9 group is a rare type of anthocyanin-related MYB activator and only present in a limited number of plant species. Secondly, all previously reported MYB10 genes, such as AtPAP1/AtMYB75 (Borevitz et al., 2000), MdMYB10 (Espley et al., 2007), PpMYB10.1 (Zhou et al., 2015b), and VvMYBA1 (Walker et al., 2007), show a strong activation effect on the UFGT promoter. In contrast, the peach PpMYB9 is unable to activate the PpUFGT promoter, but it shows a strong activation on the PpUGT78A2 promoter. Moreover, the PpMYB10.2 gene, a member of the MYB10 group, displays no activation effect on the PpUGT78A2 promoter. These results indicate that divergence in target genes has occurred between the MYB9 and MYB10 activators. Thirdly, two previously reported MYB10 genes, PpMYB10.1 (Rahim et al., 2014; Zhou et al., 2015b) and PpMYB10.4 (Zhou et al., 2014) are related to anthocyanin pigmentation in fruit and leaf of peach, respecitvely. Here, our study reveals that an additional MYB10 gene PpMYB10.2 is involved in peach flower coloration. This suggests that the MYB10 gene has undergone duplication and subsequent divergence in spatial expression patterns. However, PpMYB9 is exclusively expressed in flower of peach (Figure 1), and its expression profile is different from those of the MYB10 genes in peach.
Taken together, the above results suggest that the PpMYB9 gene represents a novel group of anthocyanin-related MYBs, which may have diverged in function from the MYB10 genes in plants. In addition, it is worth noting that a R2R3-MYB gene in rose, designated RhMYB10, was reported in a previous study (Lin-Wang et al., 2010). RhMYB10 is able to activate transcription of Arabidopsis DFR gene when it was co-transformed with bHLHs. RhMYB10 was classified as a MYB10 gene although it represents an outgroup of the MYB10 genes in the phylogenetic tree (Lin-Wang et al., 2010). Here, our study indicates that RhMYB10 is closely related to PpMYB9, and should be grouped into the MYB9 group. This finding will be useful for both identification of the MYB9 gene in other species and pursing a comprehensive understanding of the evolutionary process of R2R3-MYB genes in plants.
Author Contributions
YH and HZ conceived and designed the experiments. HZ, QP, LL, LW, and XD performed the experiments. YH and HZ wrote the paper. JZ, AO, FR, and QJ revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by funds received from the National Natural Science Foundation of China (31601740 and 31672134) and the Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (Grant No. SAJC201327).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01557
References
- Albert N. W., Lewis D. H., Zhang H., Schwinn K. E., Jameson P. E., Davies K. M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65 771–784. 10.1111/j.1365-313X.2010.04465.x [DOI] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y. J., Blount J., Dixon R. A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393. 10.2307/3871236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24 1242–1255. 10.1105/tpc.111.095232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69 1030–1042. 10.1111/j.1365-313X.2011.04853.x [DOI] [PubMed] [Google Scholar]
- Cheng J., Liao L., Zhou H., Gu C., Wang L., Han Y. P. (2015). A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J. Exp. Bot. 66 7227–7239. 10.1093/jxb/erv419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Wei G., Zhou H., Gu C., Vimolmangkang S., Liao L., et al. (2014). Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 166 1044–1058. 10.1104/pp.114.246876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L. W., Zhou X., Burke S., Wu X., Prior R. L., Li L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154 1470–1480. 10.1104/pp.110.164160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley R. V., Hellens R. P., Putterill J., Stevenson D. E., Kutty-Amma S., Allan A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 414–427. 10.1111/j.1365-313X.2006.02964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G. (1991). Flavonoids as flower pigments: the formation of the natural spectrum and its extension by genetic engineering. Plant Breed 106 1–26. 10.1111/j.1439-0523.1991.tb00474.x [DOI] [Google Scholar]
- Fraser L. G., Seal A. G., Montefiori M., McGhie T. K., Tsang G. K., Datson P. M., et al. (2013). An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genomics 14:28 10.1186/1471-2164-14-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57 761–780. 10.1146/annurev.arplant.57.032905.105248 [DOI] [PubMed] [Google Scholar]
- Han Y., Vimolmangkang S., Soria-Guerra R. E., Rosales-Mendoza S., Zheng D., Lygin A. V., et al. (2010). Ectopic expression of apple F3’H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 153 806–820. 10.1104/pp.109.152801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. P., Vimolmangkang S., Soria-Guerra R. E., Korban S. S. (2012). Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 63 2437–2447. 10.1093/jxb/err415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 13 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppel S. C., Jaffé F. W., Takos A. M., Schellmann S., Rausch T., Walker A. R., et al. (2013). Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 82 457–471. 10.1007/s11103-013-0074-8 [DOI] [PubMed] [Google Scholar]
- Howe E. A., Sinha R., Schlauch D., Quackenbush J. (2011). RNA-Seq analysis in MeV. Bioinformatics 27 3209–3210. 10.1093/bioinformatics/btr490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. C., Chen Y. Y., Tsai W. C., Chen W. H., Chen H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168 175–191. 10.1104/pp.114.254599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J. M., Chia L. S., Goh N. K., Chia T. F., Brouillard R. (2003). Analysis and biological activities of anthocyanins. Phytochemistry 64 923–933. 10.1016/S0031-9422(03)00438-2 [DOI] [PubMed] [Google Scholar]
- Liao L., Vimolmangkang S., Wei C., Zhou Z., Korban S. S., Han Y. (2015). Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 6:243 10.3389/fpls.2015.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50 10.1186/1471-2229-10-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., Van Der Woude K., Souer E., de Vetten N., Mol J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11 1433–1444. 10.2307/3870973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J. F., Va K., Mol J. N., Koes R. (1998). Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13 475–488. 10.1046/j.1365-313X.1998.00046.x [DOI] [PubMed] [Google Scholar]
- Rahim M. A., Busatto N., Trainotti L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240 913–929. 10.1007/s00425-014-2078-2 [DOI] [PubMed] [Google Scholar]
- Ramsay N. A., Glover B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Ravaglia D., Espley R. V., Henry-Kirk R. A., Andreotti C., Ziosi V., Hellens R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:68 10.1186/1471-2229-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18 831–851. 10.1105/tpc.105.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sasaki N., Ohmiya A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54 733–749. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- Tong Z., Gao Z., Wang F., Zhou J., Zhang Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:71 10.1186/1471-2199-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu C., Katayama H., Makino I., Inagaki A., Arakawa O., Martin C. (2014). Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J. Exp. Bot. 65 1081–1094. 10.1093/jxb/ert456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., Abbott A. G., Scalabrin S., Jung S., Shu S., Marroni F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45 487–494. 10.1038/ng.2586 [DOI] [PubMed] [Google Scholar]
- Walker A. R., Lee E., Bogs J., Mcdavid D. A., Thomas M. R., Robinson S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49 772–785. 10.1111/j.1365-313X.2006.02997.x [DOI] [PubMed] [Google Scholar]
- Wang L., Zhao S., Gu C., Zhou Y., Zhou H., Ma J., et al. (2013). Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol. Biol. 83 365–377. 10.1007/s11103-013-0093-5 [DOI] [PubMed] [Google Scholar]
- Yamagishi M., Toda S., Tasaki K. (2014). The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 201 1009–1020. 10.1111/nph.12572 [DOI] [PubMed] [Google Scholar]
- Yuan Y. W., Sagawa J. M., Frost L., Vela J. P., Bradshaw H. D., Jr. (2014). Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytol. 204 1013–1027. 10.1111/nph.12968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Liao L., Gu C., Lu Z., Allan A. C., et al. (2015a). Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front. Plant Sci. 6:908 10.3389/fpls.2015.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Wang H., Gu C., Dare A. P., Espley R. V., et al. (2015b). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou H., Lin-Wang K., Vimolmangkang S., Espley R. V., Wang L., et al. (2014). Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 14:388 10.1186/s12870-014-0388-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.