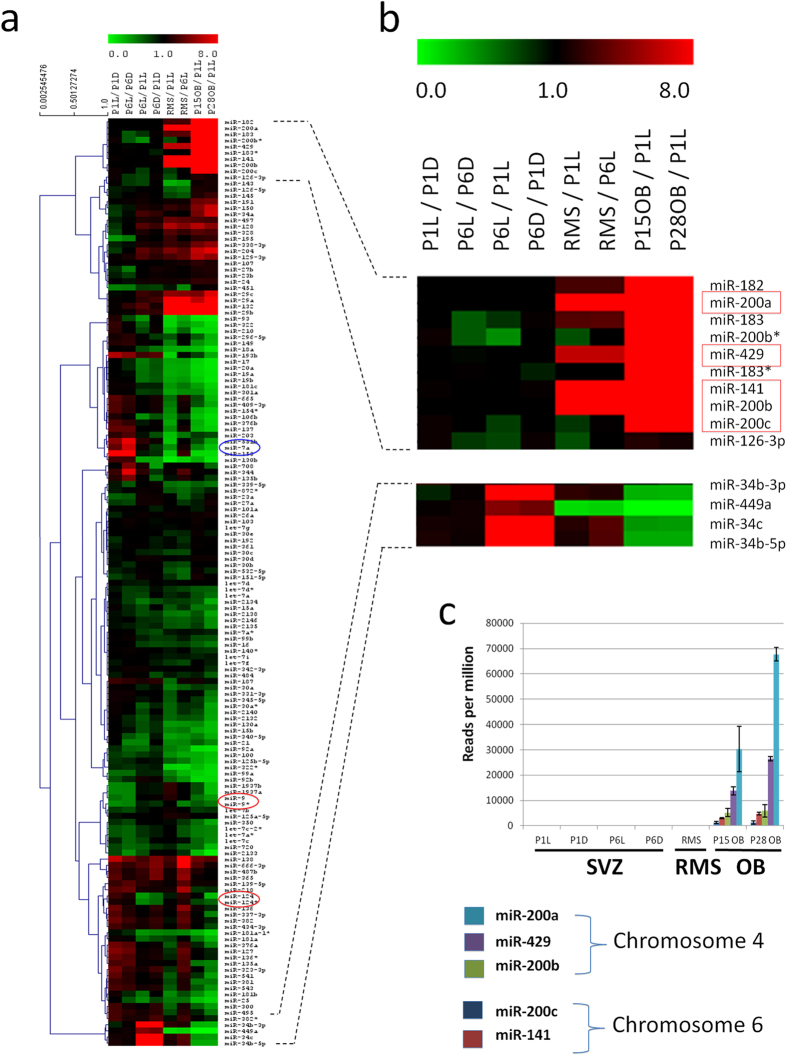
Figure 1. miRNome profiling by deep-sequencing.
(a) Heatmap showing the results of deep sequencing analyses. Only microRNAs representing more than 0.1% of total microRNAs in at least one tissue are shown. Number of reads for a microRNA in a given tissue was obtained by averaging the different sample repetitions. Columns describe the ratios between tissues selected for comparison. Proximity of vertical position indicates the similarity of expression profile of detected microRNAs and was determined using the MeV application60. (b) Close-up of specific regions of the heat map highlighting the miR-200-family a group of microRNAs preferentially expressed in the OB and the miR-34 family that is induced during ciliogenesis. miR-34a does not cluster in the heatmap due to strong expression in OB glia. (c) Histogram representing the absolute number of reads per tissue obtained in the deep sequencing analyzis for each member of the miR-200 family. All miR-200 family members are exclusively expressed in the OB but not in the stem cell or migratory compartments.