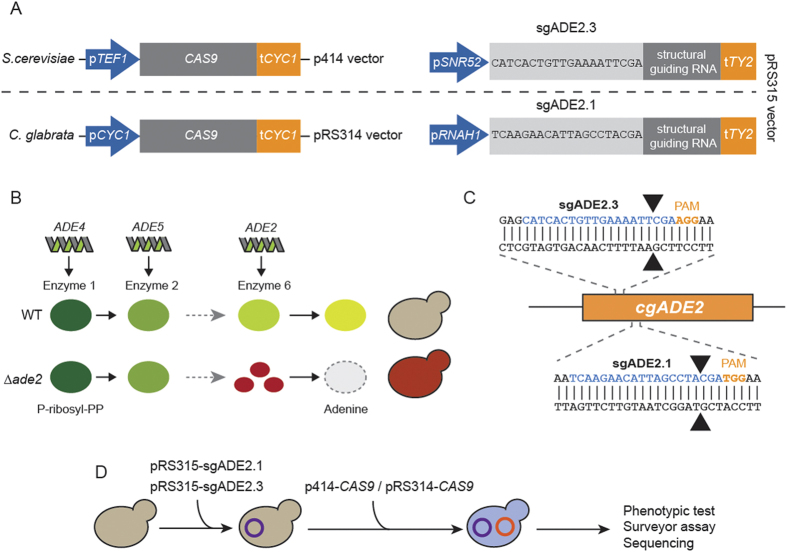
Figure 1. Design of CRISPR-Cas9 in C. glabrata and schematic of ADE2 disruption.
(A) CAS9 expression was done either with the p414 vector engineered by DiCarlo and colleagues (39) under the S. cerevisiae TEF1 promoter (pTEF1) or with the pRS314 under the C. glabrata CYC1 promoter (pCYC1). sgRNAs were cloned in a pRS315 and expressed either under the RNA Pol III promoters of S. cerevisiae SNR52 (pSNR52) or C. glabrata RNAH1 (pRNAH1). (B) Simplified scheme of adenine biosynthesis. Cells deprived of ADE2 display a red phenotype due to the accumulation of a red pigment. (C) Two target sequences localized in 5′ of the ADE2 were chosen for gene disruption: sgADE2.1 and sgADE2.3. Arrowheads indicate Cas9 cleavage sites. (D) Workflow of the sequential transformations done for sgRNA and CAS9 expression in C. glabrata and description of the following experiments.