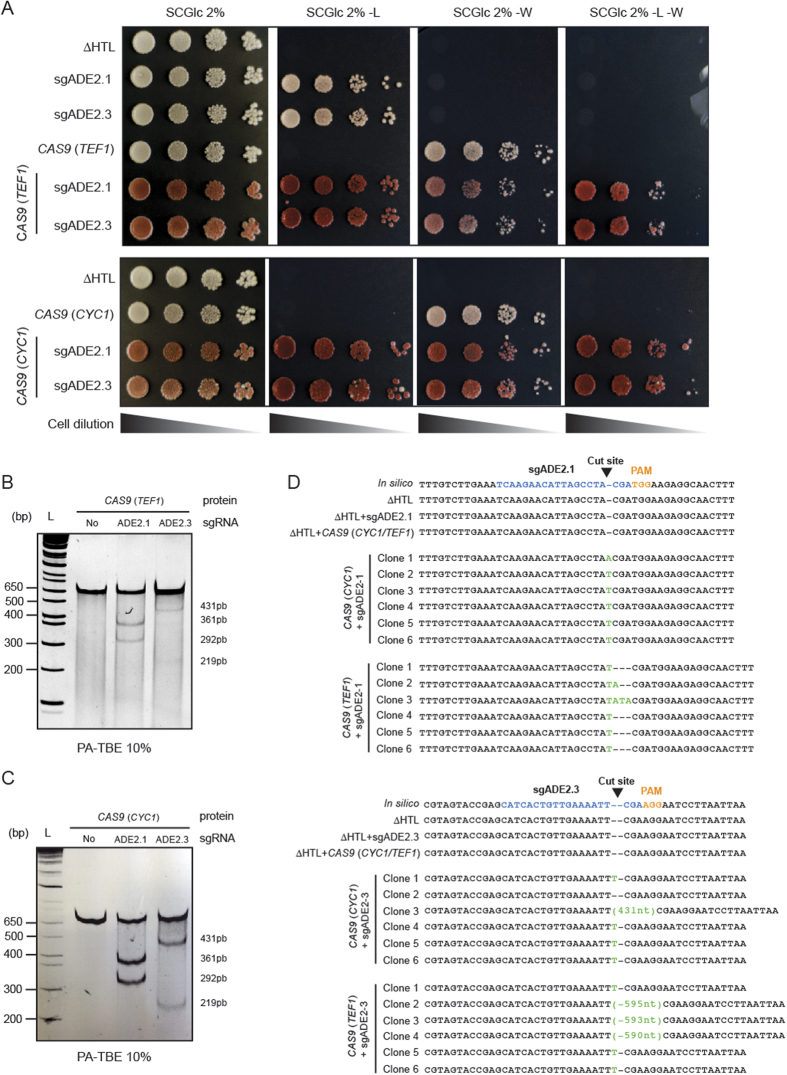
Figure 2. Efficient CRISPR-Cas9 gene disruption of the ADE2 locus.
(A) Drop test of ∆HTL, ∆HTL + sgADE2.1 or sgADE2.3, ∆HTL + CAS9 (TEF1 or CYC1), ∆HTL + sgADE2.1 or sgADE2.3 + CAS9 (TEF1 or CYC1) on SCGlc 2% and SCGlc 2% without leucine (-L) or tryptophan (-W) or both -L-W. Plates were incubated 2 days at 30 °C. (B,C) Indels in ADE2 cells were monitored by Surveyor assay in ∆HTL strain compared to the ∆HTL + CAS9 (CYC1), ∆HTL + sgADE2.1 + CAS9 (CYC1) or ∆HTL + sgADE2.3 + CAS9 (CYC1). (D) Events leading to ADE2 disruption were monitored by sequencing each strain and by comparing to the in silico constructs. Insertions and deletions are shown in green. For each deleted strain, 6 different clones were tested.