Fig. 4.
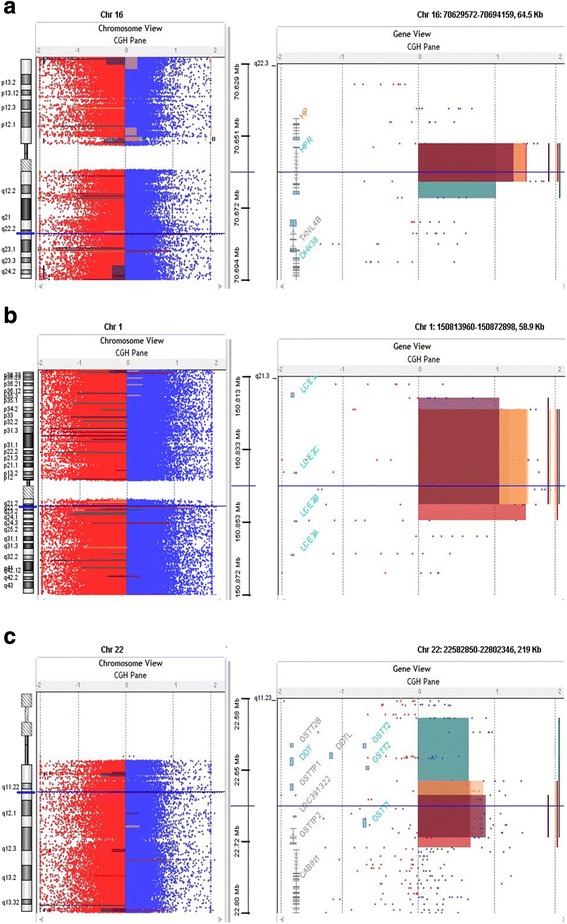
a Array-CGH profiles analysis using Agilent CytoGenomic Analytics software (V.3.0.6.6). Zero value indicates equal fluorescence intensity ratio between the sample and reference. Copy number losses shifted the ratio toward left (red) whereas copy number gains towards the right side (blue). CNVs gain at chromosome 1 starts from 150,819,879–150,819,938 cytoband 1q21.3 in the four affected members of the family and the potential gene in this region is LCE3C. b Our results showed the gain at chromosome 16 that starts from 70,647,078–70,669,681 cytoband 16q22.2 in our all four affected members of the family and the potential gene in this region is HPR gene. c Our results showed gain at chromosome 22 starts 22,686,690–22,735,300 cytoband 22q11.23 and the potential gene cluster in this region are GSTT1, GSTTP2, GSTT2B, GSTT2, DDT, and DDTL in all affected members of the family
