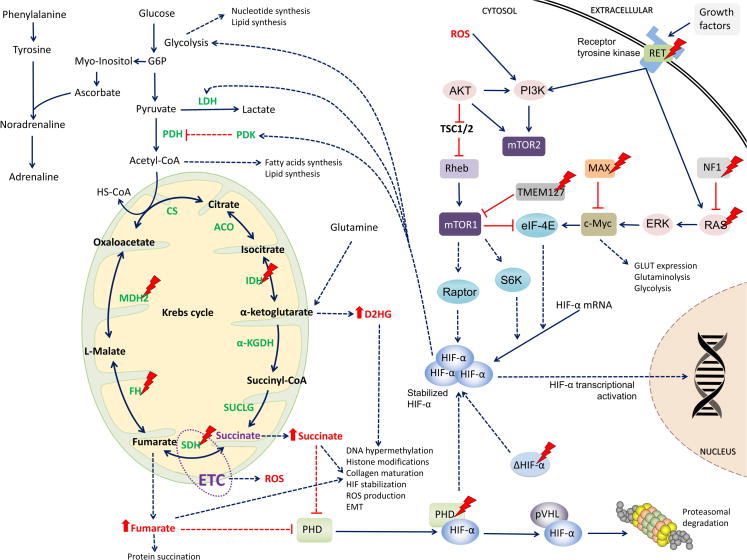
Figure 2. Metabolic changes in PHEO/PGL.
Schematic representation of mitochondrial genes as well as others involved in PHEO/PGL development with emphasis to the Krebs cycle enzymes, as explained in the text. Dotted arrows represent changes resulting from mutations in certain proteins. Actual treatment targets are explained in Table 2 and Supplementary Table S1.
Abbreviations: α-KGDH, alpha-ketoglutarate dehydrogenase; ACO, aconitase; Akt, RAC-alpha serine/threonine-protein kinase; CoA, coenzyme A; CS, citrate synthase; c-Myc, Myc proto oncogene; eIF-4E, eukaryotic translation initiation factor 4E; ERK, mitogen-activated protein kinase 2; ETC, electron transport chain; FH, fumarate hydratase; HIF-α, hypoxia-inducible factor alpha; HK, hexokinase; HS-CoA, Coenzyme A; IDH, isocitrate dehydrogenase; LDH, lactate dehydrogenase; MAX, myc-associated factor X; mTORC1, mammalian target of rapamycin complex 1; mTORC2, mammalian target of rapamycin complex 2; MDH2, malate dehydrogenase 2; NF1, neurofibromin 1; PDH, pyruvate dehydrogenase; PHD, prolyl hydroxylase domain protein; PI3K, phosphoinositide 3-kinase; pVHL, von Hippel-Lindau protein; Raptor, regulatory associated protein of mTOR; RAS, rat sarcoma oncogene; RET, rearranged during transfection proto-oncogene; Rheb, RAS homolog enriched in brain; ROS, reactive oxygen species; S6K, S6 kinase; SDH, succinate dehydrogenase; SUCLG, succinyl-CoA synthetase; TMEM127, transmembrane protein 127; TSC1/2, tuberous sclerosis complex 1/2