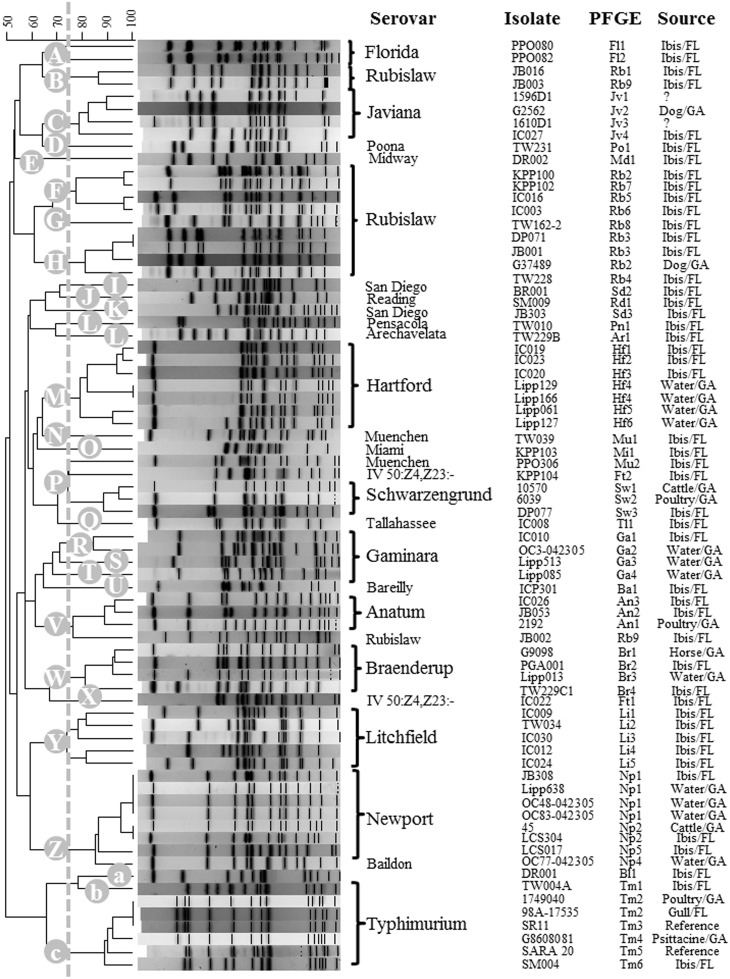
Fig 3. Cluter analysis of Salmonella.
Cluster analysis of Salmonella isolated from ibises by pulsed-field gel electrophoresis (PFGE) patterns generated with the restriction enzyme XbaI. Tiff images of Salmonella PFGE patterns were compared using DNA pattern recognition software, BioNumerics (Applied Maths, Austin, TX). Level of similarity was calculated using the band-based Dice similarity coefficient, and clustering of samples was performed using the unweighted pair-group method with arithmetic averaging (UPGMA). Salmonella PFGE patterns, generated in this study, were compared to a BioNumerics database of PFGE entries of Salmonella isolates from water and various animal species. PFGE patterns generated for ibis isolates were compared against an in-house database of Salmonella PFGE profiles for isolates from water or other animal species. A dendrogram was generated of PFGE patterns including those from isolates with the same or similar pattern (≥75% similarity). Clusters with >75% similarities were labeled A-Z, a-c on dendrogram tree on the left.