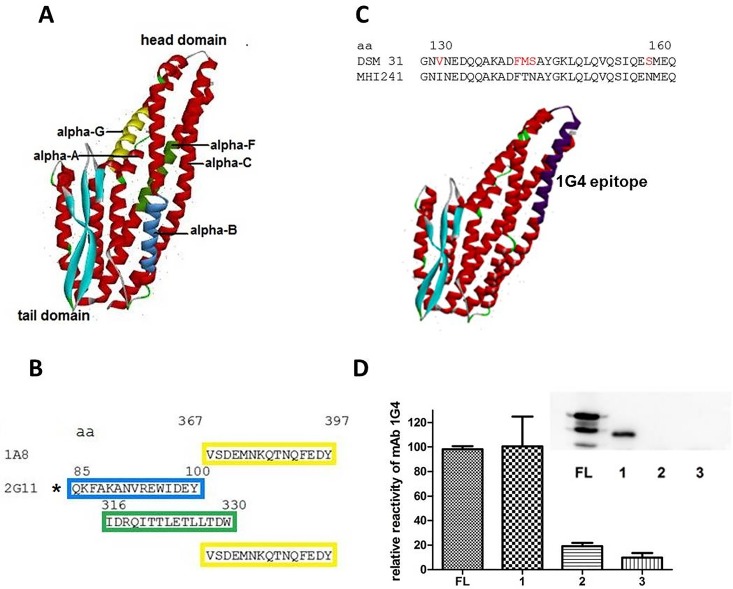
Fig 3. Epitope mapping results.
(A) NheA structure according to pdb file 4K1P with colored positions of the epitope mapping results. (B) amino acid (aa) numbers and sequences of epitopes for 1A8 (single epitope) and 2G11 (three potential binding sites the C-terminal one being identical to that of 1A8). The frame colors correspond to the colored structures in (A).The asterisk marks the likeliest epitope according to the results presented in Table 4. (C) Aa exchanges in MHI 241 compared to the type strain DSM 31 between positions 130 and 162 and location of the putative 1G4 binding site (dark purple), (D) 1G4 reactivity in indirect EIA with N-terminally deleted rNheA (FL—full length NheA, bar 1 – rNheA -109 aa, bar 2 – rNheA -164 aa, bar 3 – rNheA -185 aa. Means and SD in the depiction represent triplicate measurements. The insert shows the corresponding Western-blot results performed with mAb 1G4 and rabbit anti-mouse-HRP conjugate for detection of the recombinantly expressed NheA and its fragments.