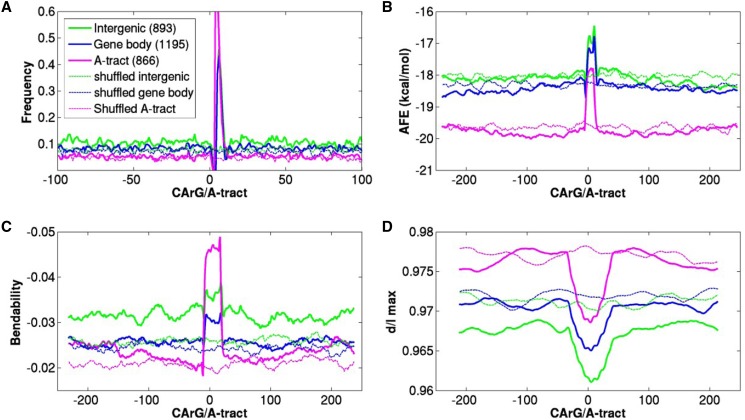
Figure 4.
Structural properties of 2954 OsMADS1-bound sequences in three data sets (intergenic, gene body, and A-tract). Sequences were aligned by positioning the CArG or A-tract motif at 0. Structural properties for sequences spanning 250 bp upstream and 250 downstream were calculated. Profiles for shuffled sequences for each data set are presented as controls in the same color as the respective OsMADS1-bound data set, but with a dashed line. A, The frequency of occurrence of A-tracts in all data sets shown only for the −100 to +100 region for clarity. B, DNA double helix stability was represented by AFE in kcal/mol. Smaller values indicate greater stability. C, Bendability or flexibility was measured by predicting DNaseI sensitivity. Smaller values suggest less bendability. D, Distribution of curvature around the motif was calculated by using BMHT dinucleotide parameters for all three data sets and their randomized controls. Smaller values indicate higher curvature.