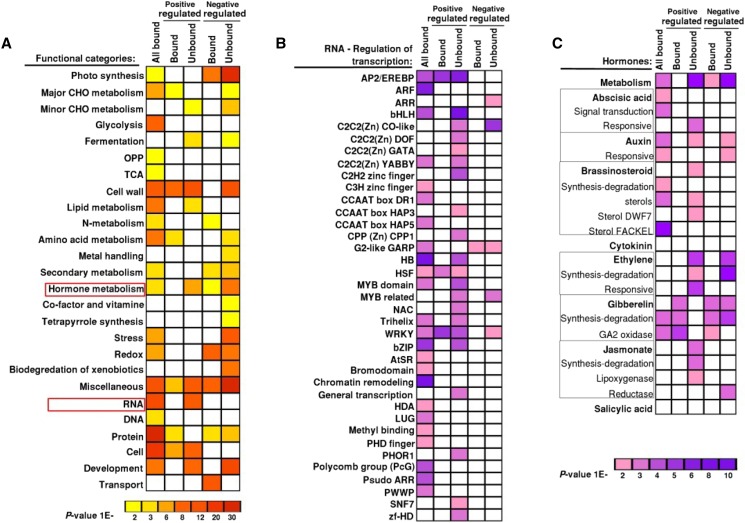
Figure 6.
Functional categories of OsMADS1-bound genes. GO term enrichment for biological processes is shown in column “All Bound” for the 3112 OsMADS1-bound gene-associated ChIP-seq data set. These classified genes are juxtaposed with GO-term-classified differentially expressed genes. Both positively as well as negatively regulated gene data sets were derived based on differential expression in 2 to 20 mm OsMADS1 knockdown versus wild-type panicles (Khanday et al., 2013). Functional classifications of bound and unbound differentially expressed genes are presented in separate columns. A, All main biological functional categories in each data set are shaded from yellow to red in the order of increasing significance. B, Enrichment for various functional families within the biological process RNA regulation of transcription. C, Representation of functional groups within hormone signaling and metabolism are shaded from pink to purple based on their increasing significance.