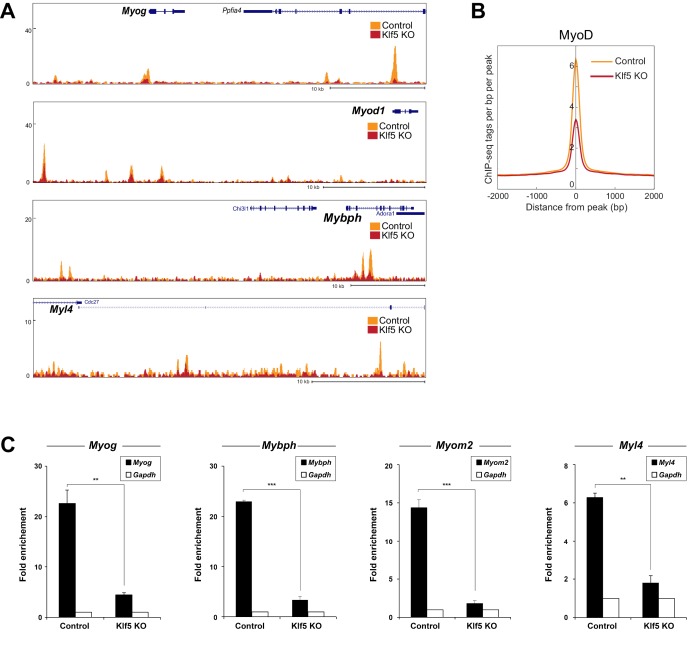
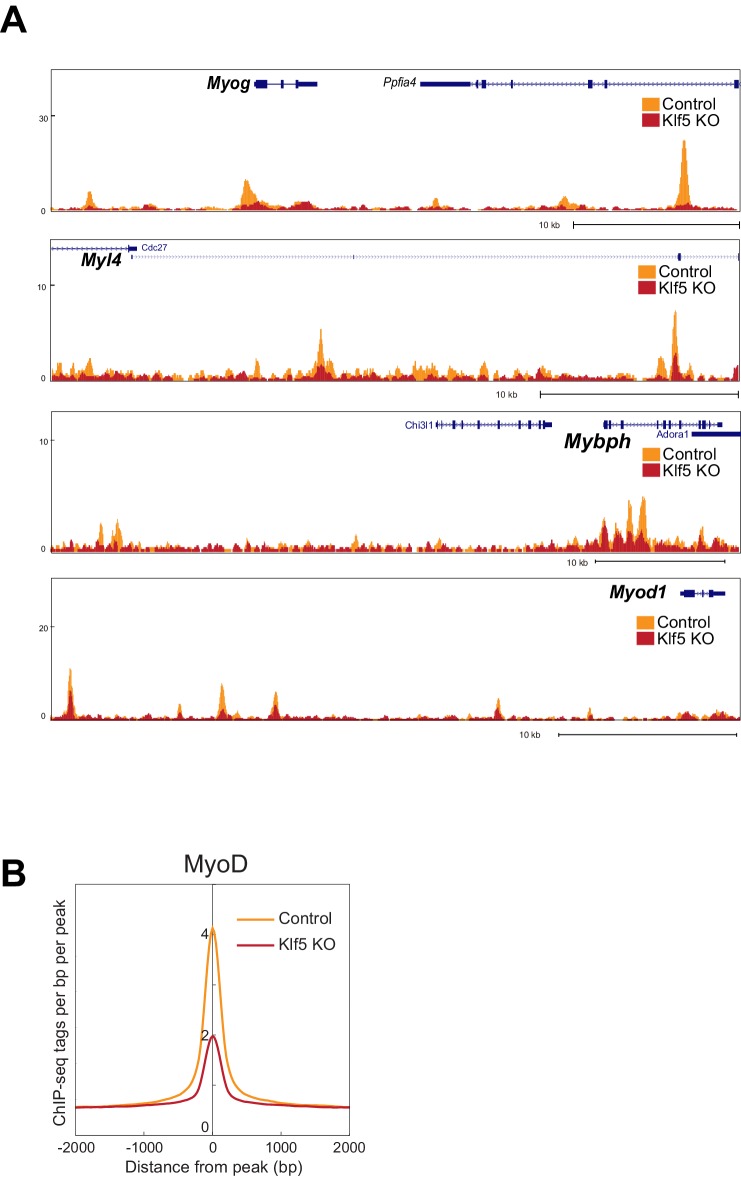
Figure 6. MyoD function is inhibited in Klf5-null C2C12 myoblasts.
(A) UCSC genome browser images illustrating normalized tag counts for MyoD at Myod1, Myog, Myl4 and Mybph gene loci in GFP-targeted control (orange) or Klf5-null (red) C2C12 myotubes differentiated for 3 days. (B) Distribution of MyoD tag densities in the vicinity of MyoD-bound enhancers in the GFP-targeted control or Klf5-null C2C12 myotubes differentiated for 3 days. (C) Comparison of MyoD recruitment at E-box containing enhancers of the Myog, Mybph, Myom2 and Myl4 gene loci in the GFP-targeted control and Klf5 null C2C12 myotubes differentiated for 3 days. Data represent means ± SEM. (**p<0.01, ***p<0.001, n = 3, biological replicates).
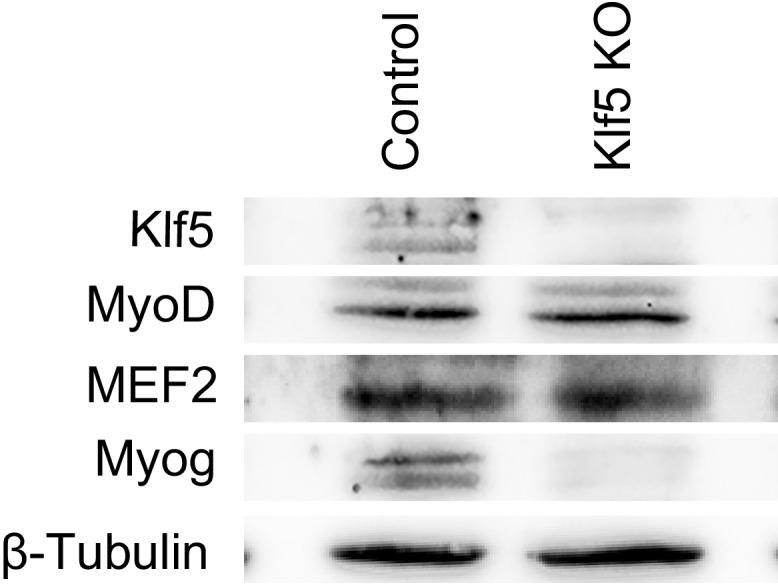
Figure 6—figure supplement 1. Levels of MyoD and Mef2 protein were not altered by Klf5 deletion.