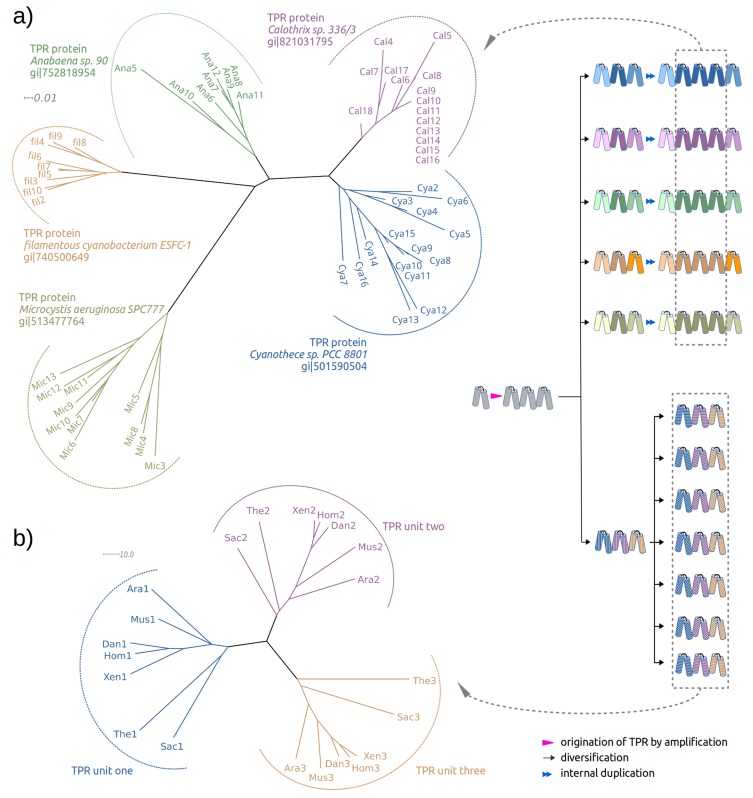
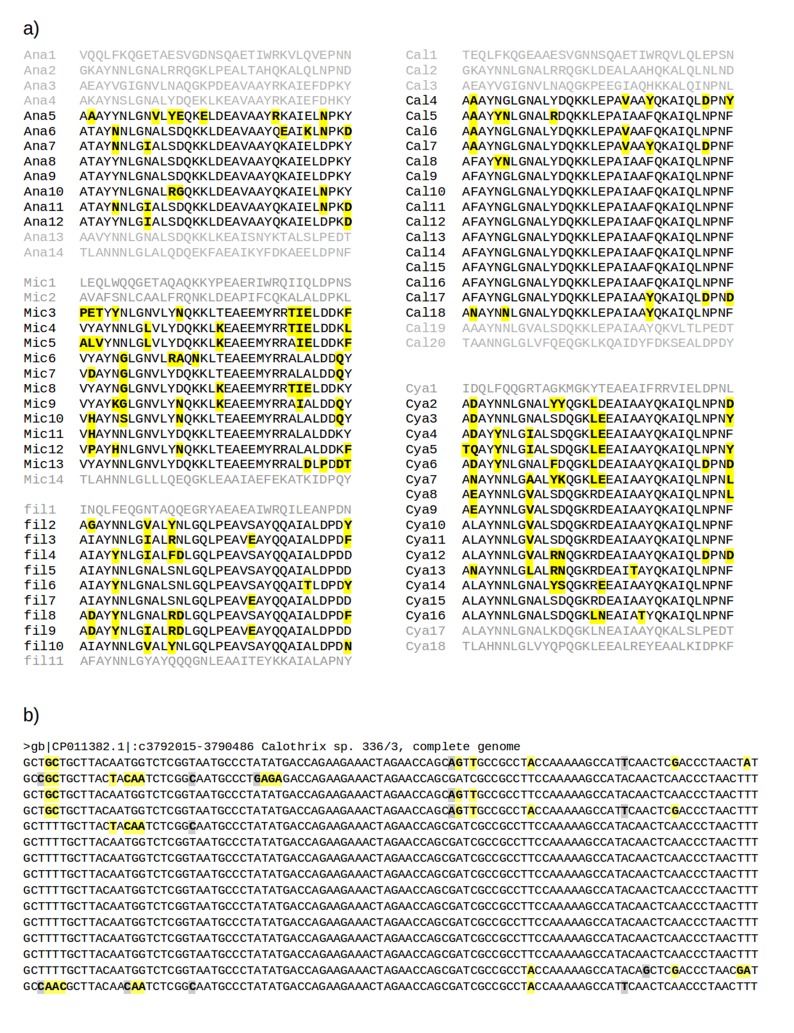
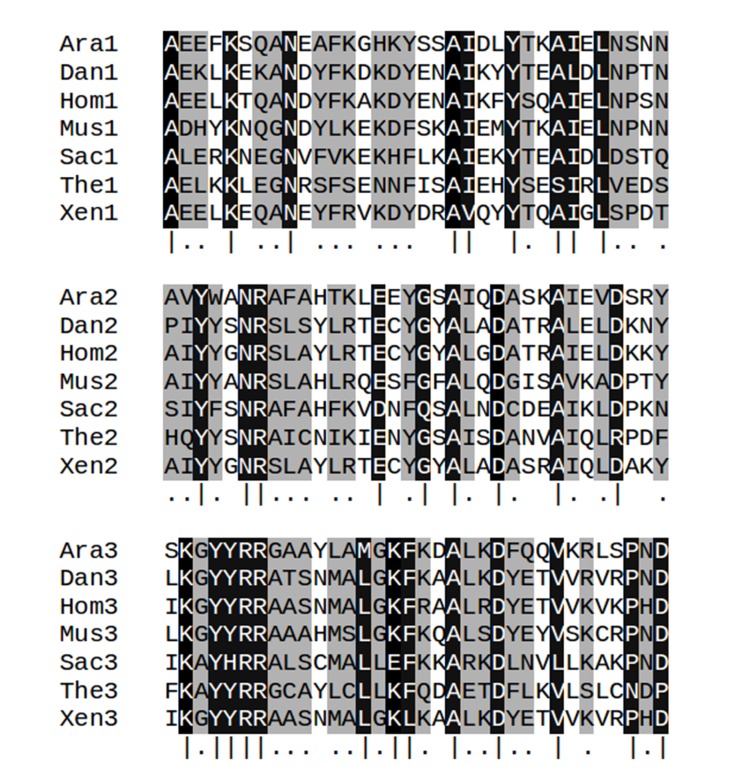
Figure 1. Two evolutionary scenarios for TPRs, illustrated by neighbor-joining phylogenetic trees.
(a) Amplification from single helical hairpin, as seen in TPR proteins from Cyanobacteria. (b) Divergent evolution of a TPR with multiple repeat units, as seen in the TPR domains of Serine/threonine-protein phosphatase 5 (Ara: Arabidopsis thaliana, Dan: Danio rerio, Hom: Homo sapiens, Mus: Musca domestica, Sac: Saccharomyces cerevisiae, The: Theileria annulata, Xen: Xenopus (Silurana) tropicalis). Since evolutionary reconstructions are subject to Occam’s razor and reflect the hypothesis with the fewest assumptions, we have postulated here one amplification event from one precursor hairpin. Our findings would however also be fully compatible with the precursor hairpin yielding a population of homologous variants, some of which were independently amplified to TPR-like folds; one or more survivors among these would have become the ancestor(s) of today’s TPR proteins. In this more complex scenario, the homology of TPR proteins, which we trace through the comparison of individual hairpins, is still given, but the TPR fold could have arisen from several independent amplifications, and not just a single one.