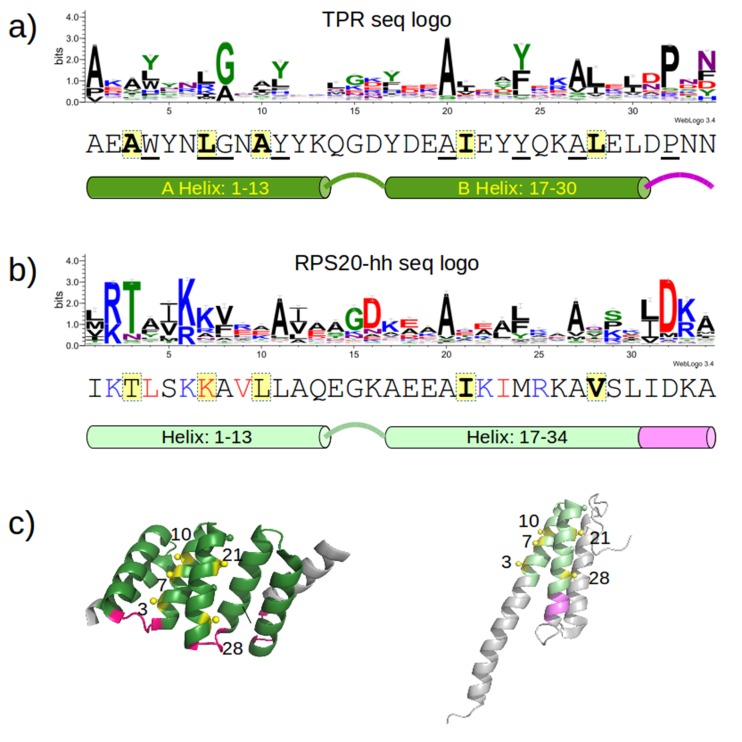
Figure 4. Sequence positions considered for optimizing the designed proteins.
(a) Sequence logo of the TPR motif. A TPR consensus sequence (Main et al., 2003b) (PDB: 1na0, chain A) and its secondary structure determined by DSSP (Kabsch and Sander, 1983) are aligned below the sequence logo. The eight TPR signature positions are underscored in the consensus sequence. The five interface positions are highlighted in yellow. (b) Sequence logo of RPS20-hh. The RPS20-hhta sequence and its predicted secondary structure using Quick2D (Biegert et al., 2006) is aligned below the sequence logo. The derived interface positions are highlighted in yellow. The four residues subjected to mutations are colored in red. The four positively charged residues selected for mutation to lower the surface charge are in blue. (c) The locations of the interface positions displayed on a TPR (left) and a RPS20 structure (right). In both structures, the interface positions are labeled and highlighted as yellow spheres. The TPR structure is CTPR3 (PDB: 1na0, chain A), which is shown as a cartoon and is colored using the same scheme as the secondary structure representation in (a). The stop helix is in gray. The RPS20 structure is from T. thermophilus (PDB: 4gkj, chain T), in which the RPS20-hh fragment is colored using the same scheme as the secondary structure representation in (b). The sequence logos were generated using WebLogo (Crooks et al., 2004). Sequences from representative proteome 75% (Chen et al., 2011) downloaded from Pfam families TPR_1 and Ribosomal_S20p were used as input to WebLogo (9338 and 972 sequences, respectively). The structures were rendered using PyMOL (Schrödinger, 2010).