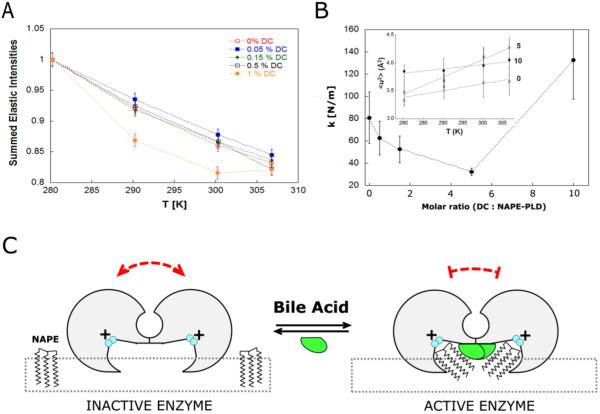
Figure 4.
The interaction of bile acids restricts NAPE-PLD structure fluctuations. (A) Elastically scattered intensity binned over the explored range of momentum transfer (Q) as a function of temperature for NAPE-PLD at different concentrations of the bile acid DCA (left panel). (B) Dependence of the force constant k (resilience) upon the molar ratio [DCA : NAPE-PLD]. Inset: temperature dependence of the mean square displacements (< u2 >) for the complex between NAPE-PLD and DCA, at molar ratio 0, 5 and 10. (C) Hypothetical mechanism of bile acid-mediated restriction of protein dynamics. Functional, structural and dynamics measurements are consistent with a model where bile acids (green) promote the assembly of inactive NAPE-PLD subunits (gray) into an active dimer, which has a reduced dynamics. The resulting lipid-protein complex can recognize the substrate NAPE at the membrane interface (square dots) to promote its hydrolysis. The positively charged binuclear zinc center of the active site is colored in cyan.