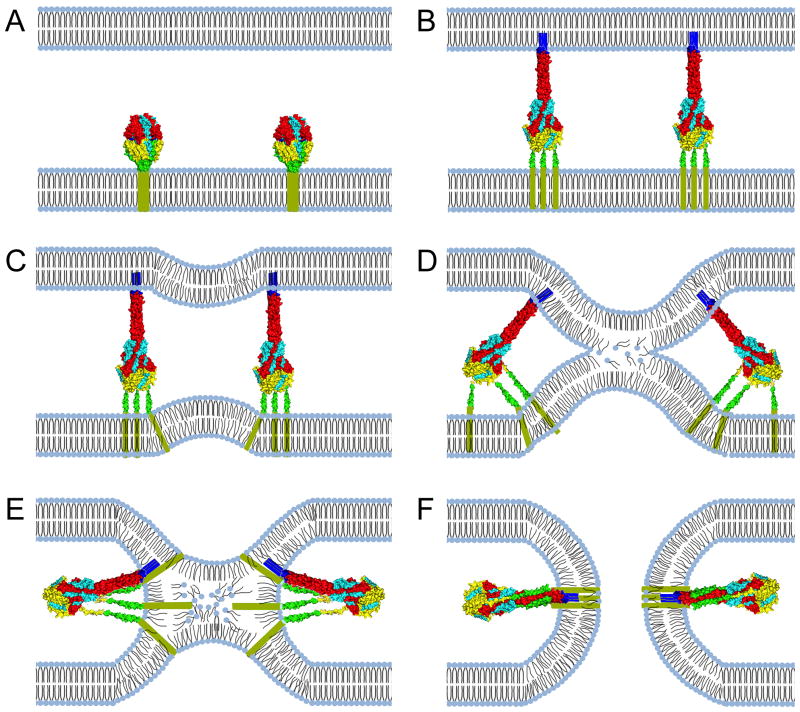
Figure 2.
Fusion pore formation through class I viral fusion proteins. (A) The F protein initially folds into a metastable prefusion conformation. (B) Once proteolytically matured and activated, the prefusion F head domain undergoes a series of major conformational changes, resulting in the assembly of an elongated trimeric coil (HR-A), which propels the fusion peptide towards the target membrane. (C) Heptad repeats proximal to the transmembrane domain (HR-B) start to fold onto the HR-A central helix during F hairpin formation, generating fusion dimples in the lipid bilayers. (D & E) Continued hairpin formation leads to the assembly of the 6HB through zippering of the HR-B domains into the grooves of the HR-A triple helix. Extreme negative curvature is introduced into the approaching bilayers, resulting in the disarray of lipids in the outer monolayers and ultimately merger of the outer layers in a hemifusion intermediate. (F) Transmembrane domains and fusion peptides are brought into close proximity, leading to the opening of a fusion pore. Creation of a productive fusion pore by paramyxovirus F proteins is not mandatorily dependent on full closure of the 6HB [100]. Images of protein structures were generated in Pymol. PDB codes: prefusion F:4MMS; postfusion F:3RRT