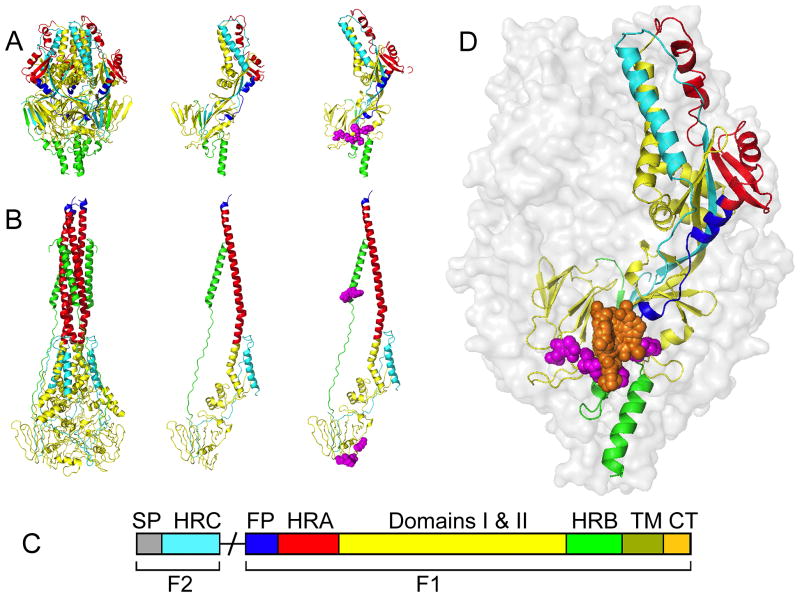
Figure 3.
Structural model of the RSV fusion protein. (A) The RSV F protein in the prefusion conformation. Shown are a ribbon model of the native prefusion F trimer (left), a single monomer (center), and a monomer with known resistance mutations (magenta) in the 392–401 and 486–489 microdomains (right). (B) The F protein in the thermodynamically stable postfusion conformation. Fields of view as described for (A). (C) Schematic of the domain architecture of the RSV fusion protein. Proteolytic maturation of newly synthesized F results in the liberation of the F1 and F2 subunits. F1 harbors the fusion peptide (FP; blue), heptad-repeat A (HR-A; red), connecting domains 1 and 2 (yellow), heptad-repeat B (HR-B; green), a transmembrane domain (TM; olive), and a short cytoplasmic or luminal tail (CP; tan). F2 remains an integral structural component of the trimer. This subunit contains the signal peptide (SP; grey) and a heptad-repeat C (HR-C; cyan) domain. (D) Schematic of the prefusion RSV F trimer overlaid with entry inhibitor binding locations (orange) and resistance hot spots (magenta spheres). For clarity, only one monomer was color-coded by domain structure as outlined in (C). Images of protein structures were generated in Pymol. PDB codes: prefusion F:4MMS; postfusion F:3RRT; Inhibitor bound F: 5EA3, 5EA4, 5EA5, 5EA6, 5EA7.