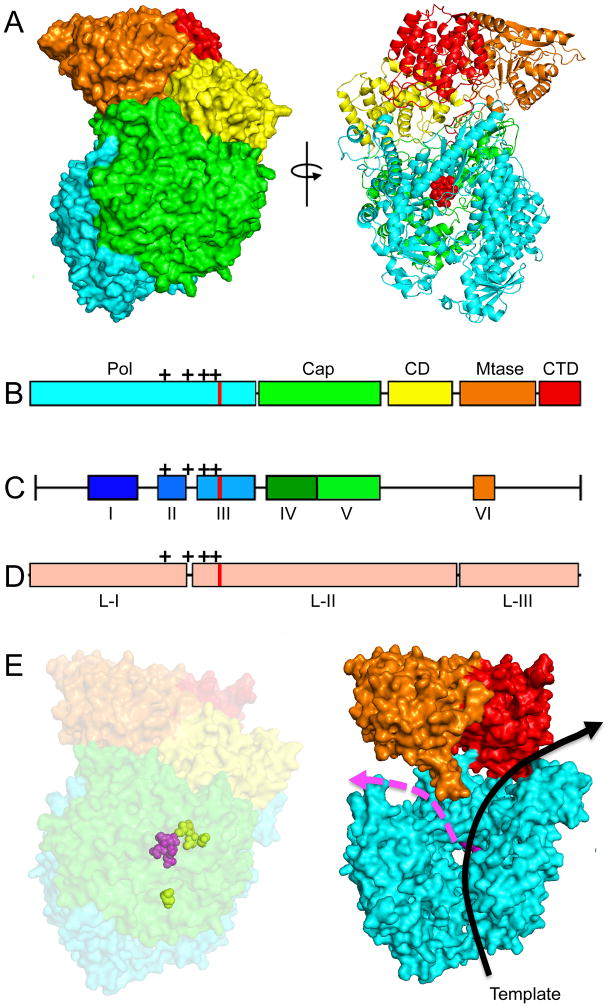
Figure 5.
Structure of a mononegavirales L protein and homology models of the RSV L protein generated based on the coordinates reported for the related VSV L structure [65]. (A) Structure of the VSV L protein shown as surface (left) and ribbon (right) models. Red spheres in the ribbon model highlight the active site for phosphodiester bond formation. (B) The VSV L structure displayed five main structural units: polymerase (cyan), capping (green), connector domain (yellow), methyltransferase (orange), and C–terminal domain (red). (C) Sequence analysis revealed six conserved regions (CR I–VI) in the L proteins. (D) Insertion analysis has shown that paramyxovirus L proteins can be further divided into three large regions (LRI-III). The location of the polymerase active site is shown in (B–D) as red lines, mutations that confer resistance to the RSV nucleoside analog inhibitor ALS-8176 are shown as black crosses. (E) Homology models of the RSV L protein based off the VSV L structure. A surface representation (left) and a model of the internal organization (right) are shown. The active site for phosphodiester bond formation and the ALS-8176 resistance mutations are highlighted as magenta and yellow spheres, respectively. Arrows indicate channels for the template (black) and newly synthesized (magenta) RNA strands. Images of protein structures were generated in Pymol; PDB code:5A22 (VSV L). Homology models were generated using SWISS-MODEL.