FIG 6.
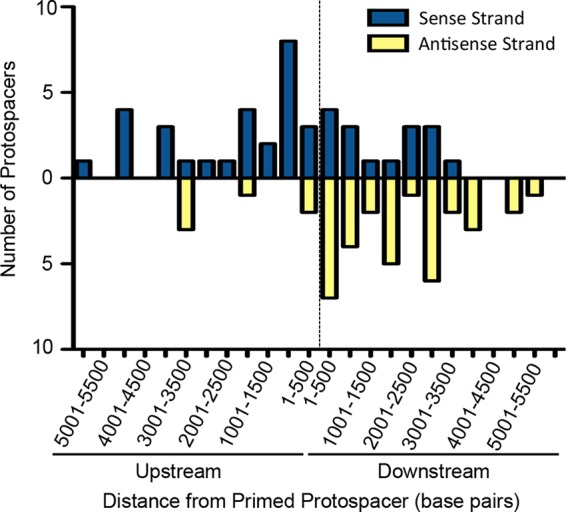
DMS3 targets of newly acquired spacers demonstrate a bias. The 83 sequenced spacers acquired by WT P. aeruginosa, when incubated with DMS3vir in the biofilm enrichment assay, within 6,000 bp of the CRISPR2 spacer 1 (CR2_sp1) target are displayed as a function of both the distance from the CR2_sp1 target and which DMS3 strand (sense or antisense) that each targets. The dashed line represents the CR2_sp1 target location, with the x axis representing the distance from the CR2_sp1 target, divided into segments of 500 bp either upstream or downstream of the CR2_sp1 target. Each bar represents the number of spacers targeting within that particular 500-bp segment of DMS3, with blue bars representing targets on the positive-sense DMS3 strand and with the yellow bars representing targets on the negative-sense DMS3 strand.
