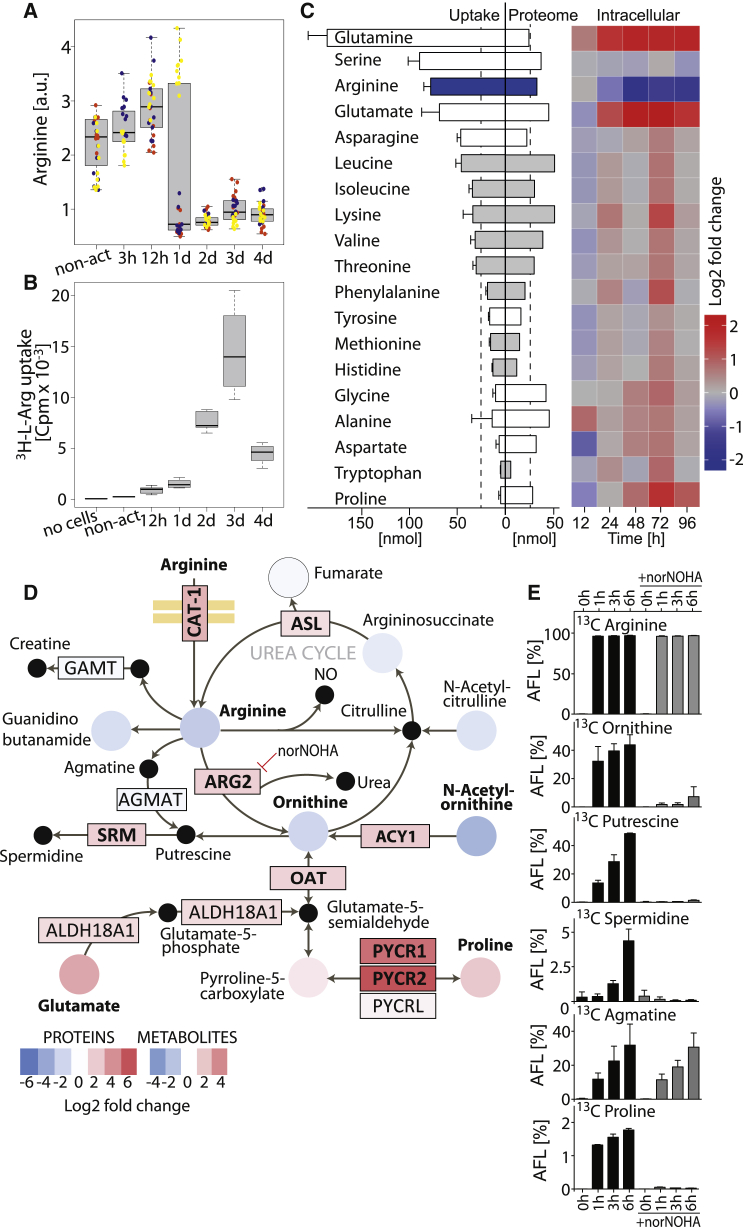
Figure 2.
L-Arginine Is Rapidly Metabolized upon Activation
(A) Intracellular abundance of L-arginine in non-activated (non-act) and activated naive CD4+ T cells (CD3 + CD28 antibodies). Boxplot, n = 30 from three donors, each in a different color.
(B) Kinetics of 3H-L-arginine uptake during a 15-min pulse. Box plot, n = 5 from three donors.
(C) Uptake, proteome incorporation and intracellular abundance of the indicated amino acids. Barplot (left): 5 × 104 cells were activated for 4 days and consumption of amino acids from medium was analyzed. Essential amino acids are in gray; n = 4 from four donors, error bars represent SEM. Barplot (center): proteome incorporation of amino acids estimated from the copy numbers of each protein. Heat map (right): intracellular amino acid abundance relative to naive T cells over time as determined by mass spectrometry (MS) n = 30 from three donors. Leucine and isoleucine could not be distinguished as they have the same mass.
(D) Changes in the abundance of metabolites and proteins of the arginine and proline metabolism between non-activated and 72 hr-activated CD4+ T cells. Log2 fold changes of proteins and metabolites are color-coded. Significant changes are in bold (FDR = 0.05, S0 = 1 for proteins; and p < 0.05 [two-tailed unpaired Student’s t test], |Log2 fc| > 1 for metabolites). Black dots are metabolites that were not detected by MS. Only enzymes that were detected by MS are shown.
(E) Metabolic tracing of L-arginine. Ninety-six hour-activated T cells were pulsed with 13C6-L-arginine and the metabolic fate was analyzed by LC-MS/MS at different time points. AFL, apparent fractional labeling; n = 4 from two donors. 13C Citrulline was not detected. Error bars represent SEM.
For (A) and (B), upper whisker = min(max(x), Q_3 + 1.5 ∗ IQR) and lower whisker = max(min(x), Q_1 – 1.5 ∗ IQR).