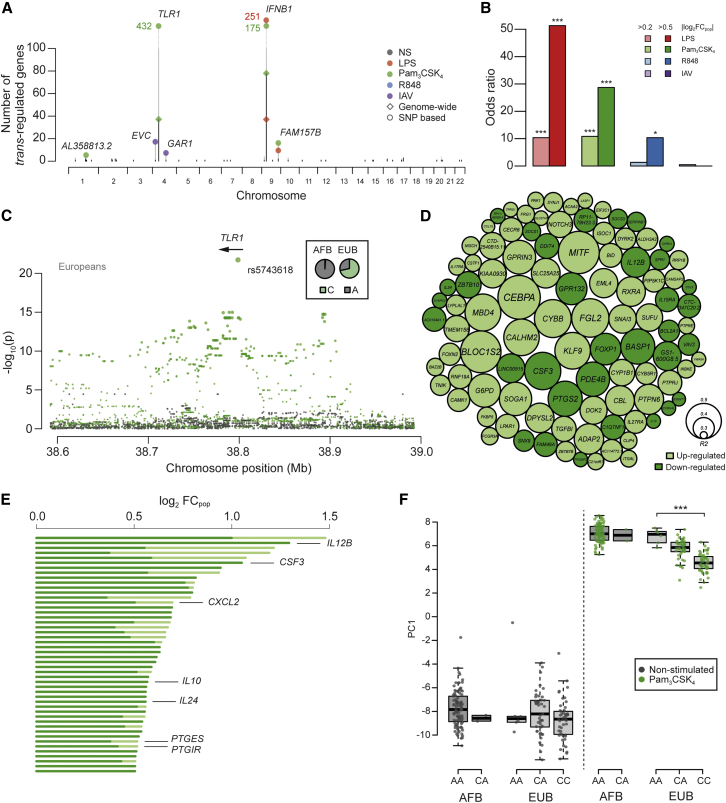
Figure 4.
Identification of Master Regulatory Loci of Immune Responses
(A) Genome-wide distribution of trans-eQTLs. For each locus, the number of associated genes identified at a genome-wide FDR of 5% or using an SNP-based Bonferroni correction is represented by black and gray bars, respectively.
(B) Enrichment of popDRGs in genes regulated by trans-eQTLs. Within each condition of stimulation, popDRGs of different strengths (light color, |log2FCpop| > 0.2; dark color, |log2FCpop| > 0.5) are represented (∗p < 0.05, ∗∗∗p < 0.001).
(C) Fine mapping of the Pam3CSK4-induced trans-eQTL at TLR1. The significance of SNP associations with the expression patterns (PC1) of the 432 trans-regulated genes is shown for basal and Pam3CSK4 conditions (gray and green dots, respectively). Only the gene overlapping the strongest trans-eQTL signal is represented.
(D) TLR1 trans-associated genes at pBonferroni < 0.05. The size of the circles reflects the proportion of the variance of gene expression explained by rs5743618, and colors indicate the direction of the change in expression associated with the derived allele. Only the 100 most significant genes are shown.
(E) Fraction of population differences in gene expression (|log2FCpop|) attributable to rs5743618 among popDRGs regulated by the TLR1 locus (in dark green). Only the tail distribution of popDRGs with the largest population differences is represented. Genes involved in the GO biological process “response to molecule of bacterial origin” are reported.
(F) Impact of the derived allele of TLR1 rs5743618 (C allele) on the expression patterns (PC1) of the 81 inflammatory genes from module 1.
See also Table S3.