Figure S5.
Genetic Structure of the Population Samples, Related to STAR Methods
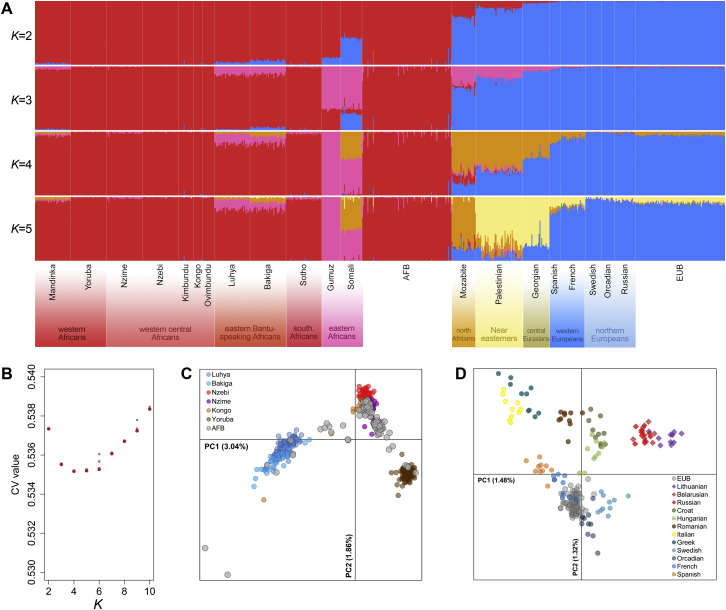
(A) Genetic ancestry of African-descent Belgians (AFB) and European-descent Belgians (AFB), estimated by ADMIXTURE. Each vertical line represents an individual genome, which is partitioned into K different genetic clusters. This analysis was performed on 229,320 independent SNPs and 789 individuals from 22 populations, including EUB and AFB, together with a selection of representative populations from sub-Saharan Africa, North Africa, the Near East and Europe (Behar et al., 2010, Patin et al., 2014). We made K vary from 2 to 10, and ran five iterations with different random seeds for each K value. The run with the lowest cross-validation error rate for each K value is shown for K = 2 to 5. (B) Cross-validation (CV) error rates of ADMIXTURE results for 5 different iterations and K prior values. Minimum CV values for each K are in red. CV values start increasing at K = 6. (C) Local genetic sub-structure in the AFB population sample, estimated by principal component analysis (PCA). This analysis was performed on 341,593 independent SNPs and 511 individuals from 7 western and central African populations (Patin et al., 2014). (D) Local genetic sub-structure in the EUB population, estimated by PCA. This analysis was performed on 182,572 independent SNPs and 220 individuals from 13 European populations (Behar et al., 2010). (C-D) PC1 and PC2 are shown, together with the proportion of variance explained.