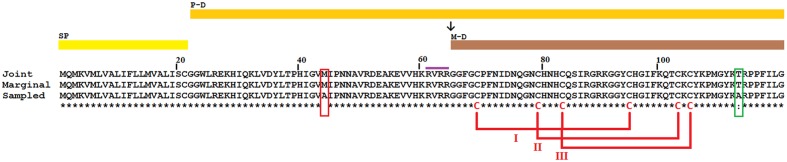
FIGURE 2.
Amino acid sequence of STiDA obtained by three reconstruction methods. Three ancestral sequence reconstruction methods were used to predict the amino acid sequence of STiDA, Joint (Pupko et al., 2000), Marginal (Yang et al., 1995), and Sample (Nielsen, 2002). The prediction by Joint and Marginal was identical. However, the prediction by Sample differed in two amino acids (red and green boxes) with that of Joint and Marginal. Conserved pattern of Cys forming putative disulfide bridges is show (numerals). The position of predicted signal peptide (SP), prodefensin (P-D), and mature defensin (M-D) are marked by colored bars. The conserved Furin recognition site (RVRR; violet bar) and the putative cleavage site (↓) are also shown.