Figure 2.
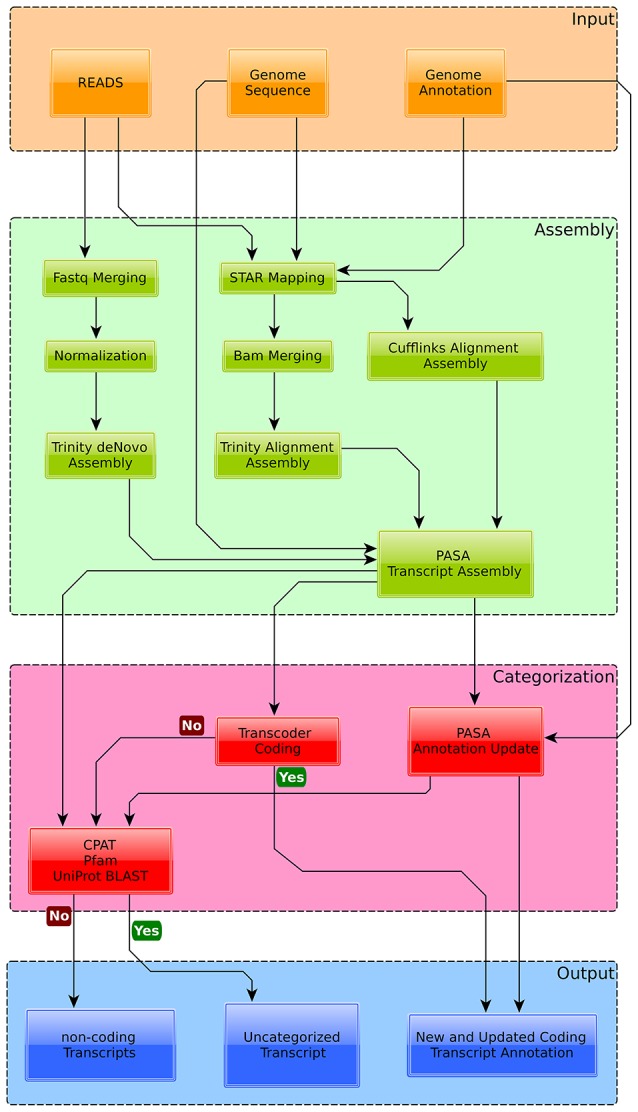
Overview of the annotation pipeline used to augment the current genome annotation of Exophiala dermatitidis. Transcripts were assembled with Trinity and Cufflinks and mapped back to the genome with PASA. Transcripts overlapping with Exophiala dermatitidis coding regions were used to find new splice variants and (re-)annotate UTRs. Transcripts that were considered coding by Transdecoder were classified as new proteins. The other transcripts were tested for their coding potential with CPAT, Pfam and by blasting them against Uniprot in order to differentiate between truly non-coding transcripts and non-coding transcripts with coding potential.
