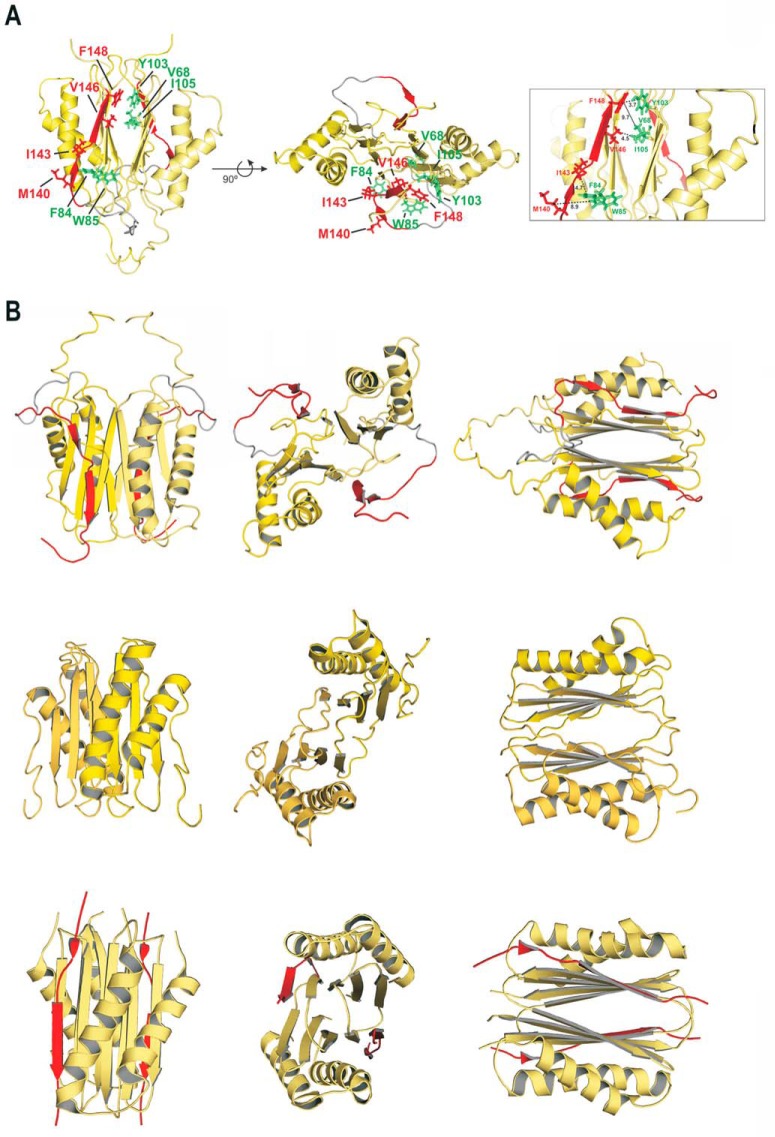
FIGURE 3.
Solution structure of human DYNLT1 self-saturated with a dynein intermediate chain polypeptide occupying the canonical binding groove. A, two different views of the calculated 3D structure of DYNLT1-DIC2 in a ribbon representation. The two chains of each DYNLT1 monomer are colored in yellow, and the DIC segment is highlighted in red. The preformed β-sheets of the monomer-monomer interaction surface become extended by swapped antiparallel β-strands, hence creating the binding surface for the incoming, kinked antiparallel β-strand of DIC. The hydrophobic contacts between the DIC segment (KLGMAKITQVDFP) and the DYNLT1 structure are also shown in a stick format. In the left view, the proposed contacts of DIC Val and Phe can be seen in the upper part of the molecule, whereas the DIC Leu, Met, and Ile are shown in the lower part of the molecule. Interacting DYNLT1 hydrophobic residues are shown in green. The right view represents a 90° rotation of the top view. The boxed panel to the right shows a close-up of the interacting residues as well as the observed distances in Å. B, structural comparison of the human, Chlamydomonas reinhardtii, and D. melanogaster DYNLT1 in complex with DIC peptides. The top three panels depict the solution structure of human DYNLT1 in complex with human DIC that we obtained using NMR spectroscopy in comparison with atomic structures of its orthologs of C. reinhardtii obtained by NMR spectroscopy (5) (Protein Data Bank code 1XDX) (three middle panels) and with the D. melanogaster crystal structure (10) (Protein Data Bank code 2PG1) (three bottom panels). Please note that the C. reinhardtii solution structure was obtained in the absence of interacting peptide.