Abstract
During 2010 many cases of discoloration in mozzarella, popularly termed as blue mozzarella, have been reported to the attention of public opinion. Causes of the alteration were bacteria belonging to the genus Pseudomonas. The strong media impact of such cases has created confusion, not only among consumers, but also among experts. In order to help improving the knowledge on microbial ecology of this microorganism a study has been set up with the collaboration of a medium-sized dairy plant producing fresh mozzarella cheese, with occasional blue discoloration, conducting surveys and sampling in the pre-operational, operational and post-operational process phase, milk before and after pasteurization, water (n=12), environmental surfaces (n=22) and the air (n=27). A shelf life test was conducted on finished products stored at different temperatures (4-8°C). Among the isolates obtained from the microbiological analysis of the samples, 60 were subjected to biomolecular tests in order to confirm the belonging to Pseudomonas genus and to get an identification at species level by the amplification and sequencing of the gyrB gene. The results of microbiological tests demonstrated the presence of microorganisms belonging to the genus Pseudomonas along the entire production lane; molecular tests showed 7 different species among the 40 isolates identified. One particular species (Pseudomonas koreensis) was isolated from blue discolored mozzarella cheese and was indicated as the most relevant for the production plant, both for the distribution along the processing chain and for the consequences on the finished product.
Key words: Pseudomonas, Dairy products, Blue discoloration
Introduction
Pseudomonas, type genus of the family Pseudomonadaceae, was originally described by Migula in 1894 and includes gram negative, aerobic, heterotrophic, with high adaptability, high metabolic versatility and ability to colonize various natural environments. The genus Pseudomonas has repeatedly undergone taxonomic revisions. Pseudomonas species were initially grouped on the basis of the results of conventional phenotypic tests. In 1973, Palleroni proposed a classification based on studies of DNA-DNA and DNA-rRNA hybridization. In this way, Pseudomonas species were classified into five clusters based on homology of ribosomal RNA, called RNA similarity groups. As time passed, the five groups turned out to be related to a wide variety of Proteobacteria (De Vos and De Ley, 1983). In the last two decades the species of Pseudomonas belonging to groups II-V have been transferred to other genera. Traditionally, the characterization of strains of Pseudomonas is performed using classical phenotypic tests such as growth on appropriate media, enzymatic reactions and microscopic analysis. However, these approaches are time consuming and are often inappropriate when examining strains belonging to closely related species. There are two methods of identification and characterization of bacteria commercially available and widely used: the systems API and Biolog. The characterization of bacterial strains using these phenotypic tests is, however, controversial, as it depends on the ability of the microorganism to grow in certain environmental conditions, and may be complicated by the presence of catabolic plasmids (Palleroni, 1986). Also, even the more discriminating phenotypic system (for example, the biotype -100 strips) is not able to resolve the species in highly heterogeneous complexes such as P. fluorescens, P. putida or P. syringae. For the identification of Pseudomonas species that produce the fluorescent pigment pyoverdin a typing method based on siderophores was proposed.
During 2010 many cases of blue discoloration in mozzarella – popularly termed as blue mozzarella – have been reported to the public opinion. The strong media impact of such cases created confusion, not only among consumers, but also among experts. On the one hand the isolation of bacteria of the genus Pseudomonas, already widely known for this type of alterations, highlights the absence of danger for the health of consumers. On the other hand, repeated seizures of products with blue discoloration and the unpredictability of the alteration have become a serious problem for producers in the dairy sector.
This study was conducted to get knowledge regarding the Pseudomonas species distribution in a dairy plant in Turin, producing different types of mozzarella and pasta filata cheese, where several cases of blue discoloration of the final product occurred and, possibly, the species causing the discoloration.
Materials and Methods
The dairy plant did have a cleaning in place system for coagulation tanks, while the sanitizing of the other equipments and plant environment was conducted by a rotation of different active ingredients and hot water. Cooling water had a supplementary chlorination.
The samples were taken at three different times, between July 2011 and February 2012. This time span was needed for the evaluation of the distribution of the organism over time. Environmental swabs were performed through the use of sponges which were rubbed on surfaces of equipment and machinery and working surfaces. For the sampling of large areas a 100 cm2 delimiter was used on 5 different points. A total of 22 environmental samples were taken.
The air sampling (n=27) was carried out in three zones of passage (forming, cooling and packaging) using sur-face-air system PBI.
Water sampling (n=12) included cooling water and water after the equipment cleaning at the end of the day. All samples (each 100 mL) were filtered under vacuum (vacuum filtration system; Millipore, Billerica, MA, USA) using 0.45 mm Millipore membrane filters. After filtration, each filter was removed aseptically from the apparatus and placed onto agar plates. Finished products (mozzarella cheese) and milk before and after heat treatment has been also sampled two times (July 2011 and November 2011). Pasteurized milk and three packages of mozzarella cheese were analyzed each time at day of production and after 7 and 15 days at two different storage temperatures (4 and 8°C). Milk before pasteurization was sampled once, during the first sampling.
All the samples were tested for the presence of Pseudomonas species (ISO/TS 11059 2009 - IDF/RM 225 2009; ISO, 2009). Bacteria were recovered onto PAB-CFC plates (Pseudomonas agar base with cetrimide, fusidin and cephaloridine) (Oxoid, Basingstoke, UK) after aerobic incubation at 25°C for 2 days. Colonies were chosen randomly and re-streaked onto Nutrient agar (NA) plates to achieve purity and stored at -80°C as 15% (w/v) glycerol stocks. These were accessed to provide fresh inocula for assays as required.
A shelf life test was conducted on the finished product as follows: three packs of mozzarella cheese were tested on the day of production and subsequently on the 7th and the 14th day at 4 and 8°C. Twenty g of mozzarella were sampled and analyzed from each pack. For species identification, a fragment of 841 base pairs (bp) of gene gyrB was amplified with UP-1E (5’-CAGGAAACAGCTATGACCAYGSNGGNGGNAARTTYRA-3’) and APRU (TGTAAAACGACGGCCAGTGCNGGRTCYTTYTCYTGRCA 5’-3’ primers (Yamamoto et al., 2000).
The polymerase chain reaction was performed in a volume of 50 L containing: 25 L of Taq PCR Master Mix Kit, 5 L for each primer, 3 L of DNA and 12 L of water. The amplification conditions were as follows: an initial denaturation step of 5 min at 94°C, 35 cycles (60 sec at 94°C, 60 sec at 58°C, 90 sec at 72°C) and a final extension of 5 min at 73°C.
The sequencing was carried out at the BMR Genomics of Padoa and the sequences obtained were analyzed using Mega 5.0. software and blasted with GenBank database. The aligned sequences were then correlated in a phylogenetic dendrogram.
Results
Milk sampled before the pasteurization process was positive for the presence of Pseudomonas spp.; isolates from this sample demonstrated the presence of at least three different species (P. fragi, P. marginalis, and P. koreensis). The milk after pasteurization was constantly negative for the presence of Pseudomonas spp. after being tested at 7 and 14 days after the treatment. In Figure 1 the results of the shelf life tests on the finished product are shown.
Figure 1.
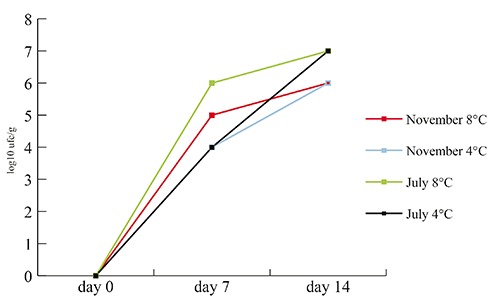
Shelf life tests of mozzarella cheese. The values (log10 ufc/mL) represent the mean of the counts of the three packages sampled in July 2011 and November 2011 after storage at 4 and 8°C.
Seventy four colonies, morphologically referable to Pseudomonas spp., were isolated during the study. Subsequently, the isolates were subjected to confirmation by amplification of a fragment of the gene gyrB, but only 60 colonies gave the expected fragment; the sequencing of the amplicons led to the identification at species level of 44 isolates. Three of these were found to be phylogenetically related, but not belonging to Pseudomonas genus (Aeromonas and Stenotrophomonas). The distribution of the different species among the other 41 isolates is shown in Figure 2. For 16 isolates, positive to the PCR, the identification remains uncertain. In Table 1 a summary of the results of the microbiological analysis is shown.
Figure 2.
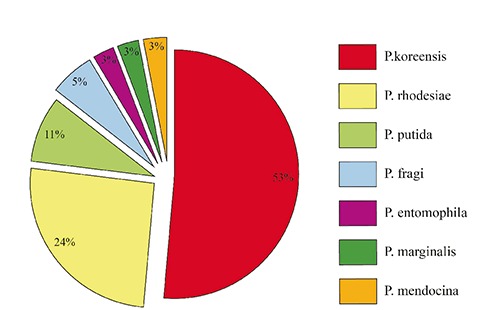
Distribution of Pseudomonas species among identified isolates (n=38).
Table 1.
Summary of microbiological results for the presence of Pseudomonas spp.
| Sample type | Total samples (n) | Positive samples | |
|---|---|---|---|
| n | % | ||
| Water | 12 | 10 | 83 |
| Air | 27 | 3 | 11 |
| Surfaces | 17 | 3 | 18 |
| Curd | 4 | 2 | 50 |
The species Pseudomonas koreensis has been found along the production chain of the dairy plant during a period of 8 months: pre-milk pasteurization, buffer surface water of the charger and the finished product, including a sampled mozzarella showing blue discoloration. On finished products only one of the species was usually present, notably P. koreensis, and, during the sampling of November 2011, P. rhodesiae, while in water and environmental samples, a wider range of species, belonging to different groups, was been found. In Figure 3 the phylogenetic dendogram derived by the analysis of the 45 sequences obtained is represented.
Figure 3.
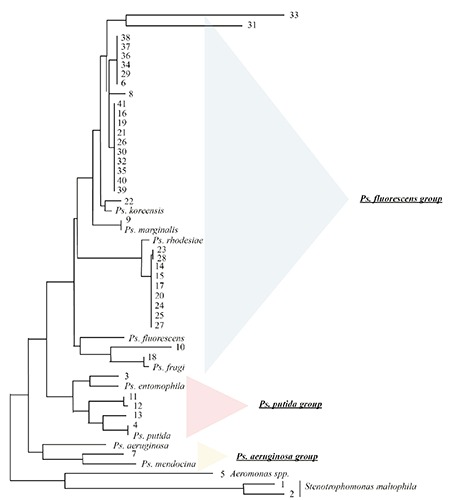
Unrooted neighbour-joining dendrogram based on gyrB sequences with Jukes-Cantor-based algorithm, 1000 bootstrap replications. Samples numbered 1-21 refer to samples collected in July 2011; 30-38 samples are those collected in November 2011; 39-41 samples are those showing blue discoloration collected in February 2012.
Discussion
P. fluorescens is a gram-negative rod with strains that are capable of producing a blue, non-diffusible pigment as well as the soluble pigment pyoverdin, which fluoresces under UV light (Palleroni, 1984). The particular spoilage investigated in this study appears to be everything but rare, and is well known that Pseudomonas spp. have been implicated in pigment-related spoilage of different foods, including yellow and blue pigments (Cantoni et al., 2003). These types of spoilage organisms most frequently contaminate products after processing (Martin et al., 2011).
Pseudomonas spp. were found in milk before the pasteurization process, but not in pasteurized milk; this leads us to the conclusion that contamination of the products has occurred during the production chain after this point, but also that raw milk can be one of the sources of contamination of the dairy plant.
The results of the shelf life tests (Figure 1) show little differences between the series of analyses at different storage temperatures, apart from a slightly steeper line for those at 8°C. The interesting fact is that quantitative analysis on the day of production gave constantly a count of <10 ufc/g, which means that a certain amount of stress (temperature during production process, chlorinated water) made the organisms non-culturable. Another possibility to take in account is the sensitivity of the analysis method: it may happen, in products like mozzarella cheese, to find negative results immediately after process steps involving thermal process (e.g. stretching), after which the contamination from environmental surfaces with a initial low bacterial charge on product occurred. Further interest should be posed to the fact that from all the different species of Pseudomonas isolated from water and environmental samples, only two (P. koreensis and P. rhodesiae) were found on the finished products and only once together. This may be due to a particular resistance of these species to stress factors during the processing or to a particular case of microbial competition. Resistance factors to sanitizers (known to be shared via plasmids among gram negative bacteria) could also play a role (Buckingham-Meyer et al., 2007).
The number of isolates identified at species level and their phylogenetic relationship is shown in Figure 3: Pseudomonas groups and subgroups clustered accordingly with previous studies (Mulet et al., 2009). The vast majority of the isolates belongs to the P. fluorescens group. The P. koreensis subgroup has different colours showing at different times: this is the only species isolated in every sampling and along the whole production chain, from raw milk to water to, eventually, the finished products, which makes this species particularly relevant for this dairy plant. This is the more so because this has been the only species found on mozzarella samples showing blue discoloration.
Conclusions
The results in our opinion highlighted the need to continue the research by inserting an enrichment step in the protocol: the results of the shelf life test, in fact, led to the supposition that viable but not culturable organisms might be present on the samples tested on the day of production, or the number of microorganisms at that time was very low, as they revealed themselves in high numbers after a week of storage. In both cases an enrichment step would overcome the problem.
The difficulties encountered to come to a complete identification of the species, finally, pushes us toward new molecular tests, shrinking the targets to the most relevant groups of Pseudomonas in dairy products (P.fluorescens and P.putida groups). The isolation of P.koreensis from blue pigmented mozzarella cheese represent a novelty, as in previous studies the causative agent was usually P. fluorescens (Martin et al., 2011; Cantoni et al., 2003).
References
- Buckingham-Meyer K, Goeres DM, Hamilton MA, 2007. Comparative evaluation of biofilm disinfectant efficacy tests. J Microbiol Meth 70:236-44. [DOI] [PubMed] [Google Scholar]
- Cantoni C, Bersani C, Dragoni I, Iacumin L, Comi G., 2003. Pseudomonas fluorescenti negli alimenti. Ind Aliment-Italy 42:609-12. [Google Scholar]
- De Vos P, De Ley J, 1983. Intra- and intergeneric similarities of Pseudomonas and Xanthomonas ribosomal ribonucleic acid cistrons. Int J Syst Bacteriol 33:487-509. [Google Scholar]
- ISO, 2009. Milk and milk products. Method for the enumeration of Pseudomonas spp. ISO Norm 11059:2009 (IDF/RM 225:2009). International Standardization Organization ed., Geneva, Switzerland. [Google Scholar]
- Martin NH, Murphy SC, Ralyea RD, Wiedmann M, Boor KJ, 2011. When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. J Dairy Sci 94:3176-83. [DOI] [PubMed] [Google Scholar]
- Mulet M, Bennasar A, Lalucat J, García-Valdés E, 2009. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol Cell Probe 23:140-7. [DOI] [PubMed] [Google Scholar]
- Palleroni NJ, 1984. Pseudomonas. Borriello SP, Murray PR, Funke G, eds. Topley and Wilson’s microbiology and microbial infections. Edward Arnold, London, pp 1591-1606. [Google Scholar]
- Palleroni NJ, 1986. Taxonomy of the pseudomonads. Sokatch JR, ed. The bacteria: the biology of pseudomonas. Academic Press, London, pp 3-20. [Google Scholar]
- Palleroni NJ, Kunisawa R, Contopoulou R, Doudoroff M, 1973. Nucleic acid homologies in the genus Pseudomonas. Int J Syst Bacteriol 23:333-9. [Google Scholar]
- Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S, 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385-94. [DOI] [PubMed] [Google Scholar]


