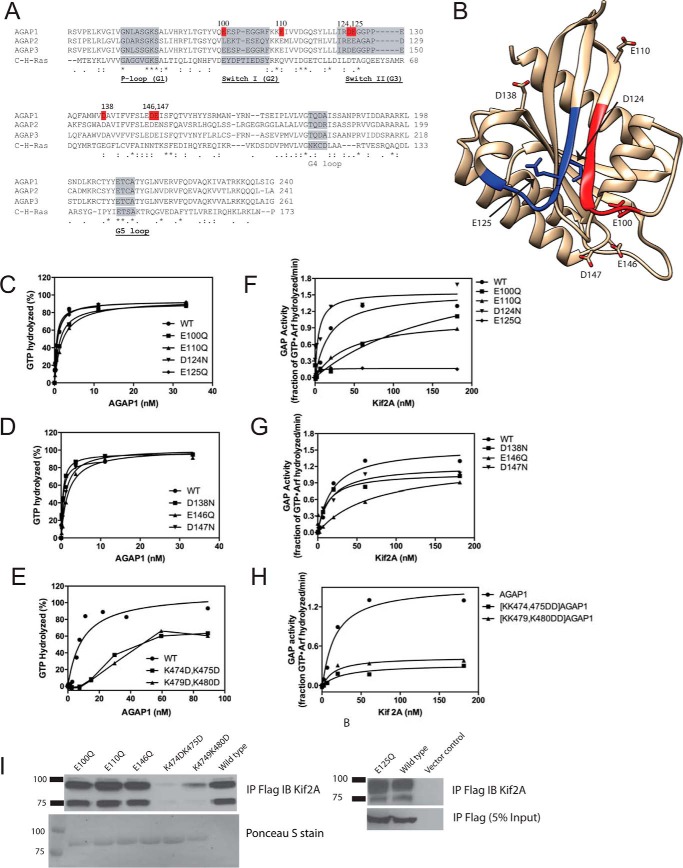
FIGURE 4.
Effect of GLD of AGAP1 on GAP activity. A, sequence alignment of AGAP GLDs with H-Ras. Accession numbers are: AGAP1, gi:51338837; AGAP2, gi:6176569; AGAP3, gi:16799069; H-Ras, gi:231061. B, structure of AGAP3 GLD. The crystal structure of AGAP3 is shown with switch 1 colored red, switch 2 colored blue, and the residues aligning with the residues mutated in AGAP1 indicated. Protein Data Bank ID: 3IHW. C–E, activity of mutant AGAPs in the absence of Kif2A. The indicated AGAP1 mutants were titrated into a GAP reaction containing LUVs (same composition as in Fig. 3A, + PtdIns(4,5)P2), and 0.5 μm myrArf1·[α32P]GTP. F–H, effect of Kif2A on the activity of AGAP1 mutants. His10-[193–531]Kif2A was titrated in a reaction containing 0.5 μm myrArf1·[α32P]GTP, 500 μm LUVs (same composition as in Fig. 3A), and 1.0 nm of wild type AGAP1 or the indicated mutants. I, binding of mutant AGAP1 to Kif2A. FLAG-tagged wild type AGAP1 or the indicated mutants were expressed in HeLa cells. Cells were lysed, and the FLAG-tagged proteins were immunoprecipitated (IP) with an anti-FLAG antibody. Kif2A was detected in the precipitates by immunoblotting (IB) using an anti-Kif2A antibody. In the left panel, immunoprecipitated FLAG-tagged protein was detected by Ponceau S staining. In the right panel, immunoprecipitated FLAG-tagged protein was detected by immunoblotting for the FLAG epitope.