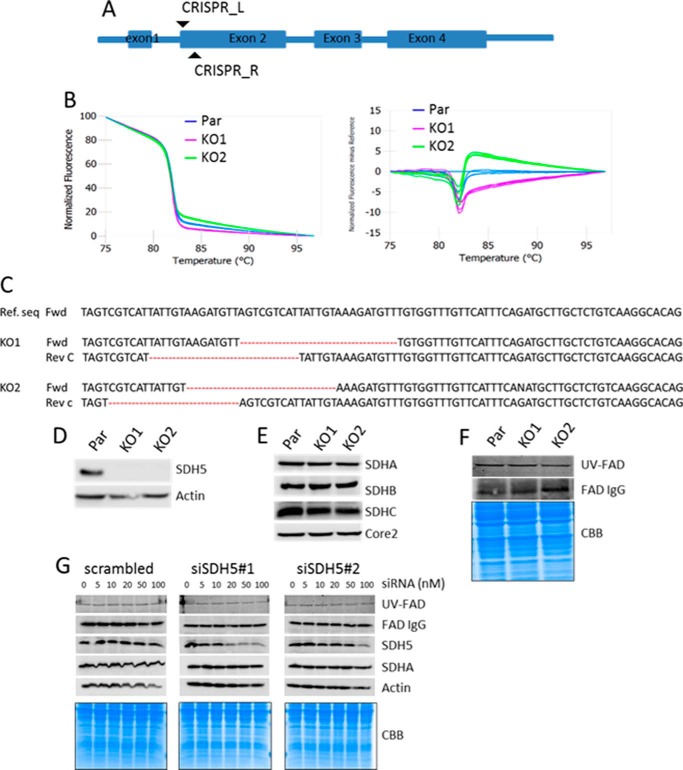
FIGURE 1.
Generation of SDHAF2 knock-out cell lines and SDHA flavination analysis. A, scheme showing SDHAF2 gene structure and sgRNA target. B, HRM curve analysis of parental cells (Par) and two selected clones. C, analysis of the nucleotide sequence spanning the SDHAF2-targeted region in the SDHAF2 gene present in the first intron and second coding exon. Reverse complement of sequence (Rev C) was used for antisense strand analyses. Ref. seq, reference sequence; Fwd, forward; Rev, reverse. D, WB demonstrating absence of SDHAF2 in KO1 and KO2 cell lines. Actin was used as a loading control. E, WB showing the steady state levels of protein subunits of CII with anti-Core2 as a loading control. F, SDHA flavination analysis using anti-FAD in WB (upper band) and fluorescent signal of FAD (middle band) from SDS-PAGE gel exposed to UV light. Coomassie Brilliant Blue R staining was used as a loading control (bottom bands). G, MDA-MB-231 cells treated with increasing concentrations of two SDHAF2 siRNAs were probed for flavination of SDHA using the UV method and probing anti-FAD IgG, as well as for the level of SDHAF2 and SDHA using WB. Coomassie Brilliant Blue R (CBB) staining and actin were used as loading control.