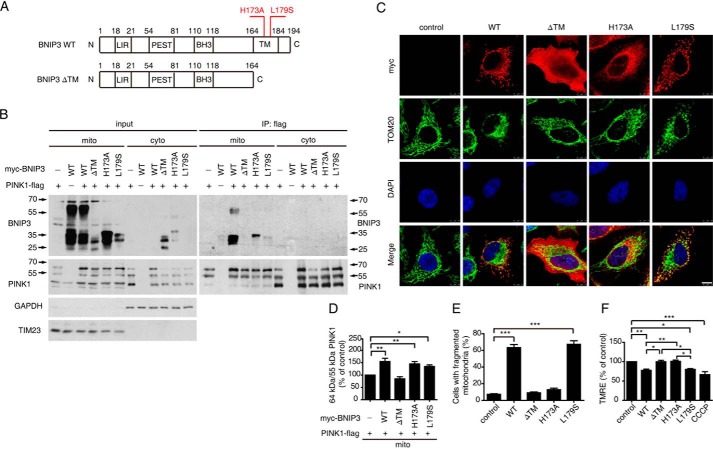
FIGURE 2.
Mitochondrial localization is essential for BNIP3 to suppress PINK1 cleavage. A, illustration of BNIP3 variants. BNIP3 ΔTM, deletion of the C-terminal transmembrane domain; BNIP3 H173A, dimerization mutant; BNIP3 L179S, depolarization mutant; LIR, LC3-interacting region; PEST, proline, glutamic acid, serine, and threonine rich sequences. B, interaction of PINK1 and BNIP3 variants. HEK293 cells expressing PINK1-FLAG and Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S) in various combinations were fractionated to separate cytoplasm (cyto) and mitochondria (mito) followed by immunoprecipitation (IP) with anti-FLAG tag (to detect PINK1) and immunoblotting with anti-BNIP3 or anti-PINK1 (right panel). Input controls are shown on the left panel. TIM23 and GAPDH, a mitochondrial marker and a cytosolic marker, respectively. C, mitochondrial morphology in cells expressing BNIP3 variants. HeLa cells expressing Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S) were immunostained with anti-Myc tag (red) or anti-TOM20 (green). Cell nuclei were labeled with DAPI (blue). Colocalization is shown on the bottom panel (Merge). Note that significant mitochondrial fragmentation is observed in cells expressing WT and L179S. Bar = 10 μm. D, 64-kDa/55-kDa PINK1 ratios affected by BNIP3 variants. Relative protein levels from the mitochondrial fraction were measured using ImageJ and analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01. E, quantitation of mitochondrial fragmentation. >300 transfected cells were counted for each experiment. The results were analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 3. ***, p < 0.001. F, mitochondrial membrane potential in cells expressing BNIP3 variants. Mitochondrial membrane potential was detected by TMRE staining followed by FACS analysis using HEK293 cells expressing Myc-tagged BNIP3 variants (WT, ΔTM, H173A, and L179S). HEK293 cells treated with CCCP (10 μm) for 2 h served as a positive control. Results were analyzed by one-way ANOVA and Dunnett's test. Error bars, S.E.; n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001.