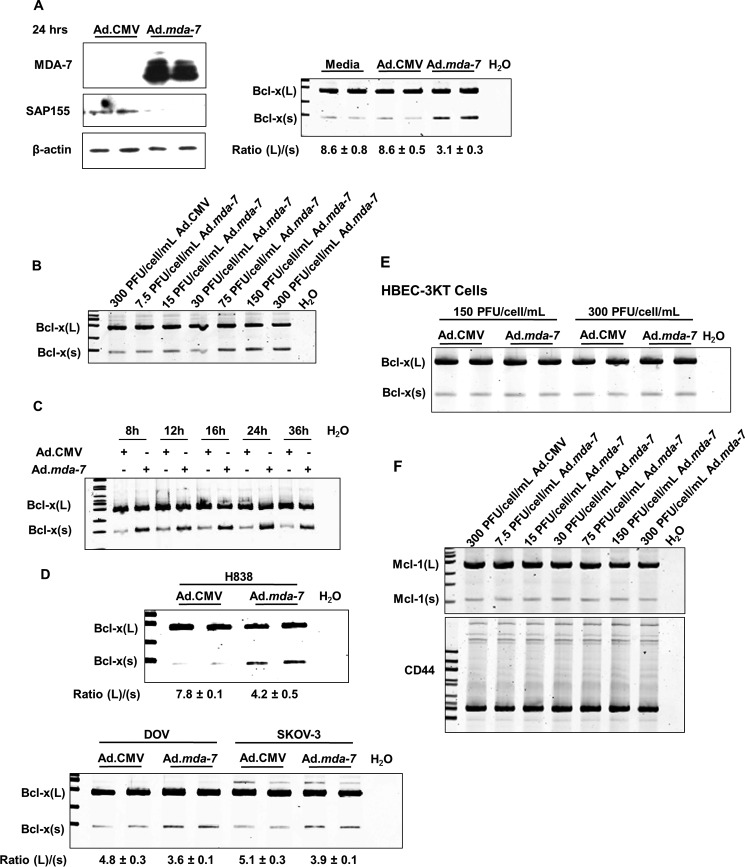
FIGURE 2.
Ad.mda-7 induces the activation of the Bcl-x(s)/proximal 5′ splice site of Bcl-x pre-mRNA. A, cells (1.2 × 105) were transduced with 150 MOI of either ad.mda-7 or Ad.CMV control virus. After 24 h, total RNA and protein were extracted. Total protein was subjected to SDS-PAGE analysis and Western immunoblotting for MDA-7, SAP155, and β-actin (left panel). Total RNA was subjected to quantitative/competitive RT-PCR analysis (competitive qRT-PCR) of Bcl-x splice variants and the corresponding Bcl-x(L)/(s) mRNA ratios (Ratio (L)/(s)). The ratio of Bcl-x(L) to Bcl-x(s) mRNA was determined by densitometric analysis of RT-PCR fragments (p < 0.01, n = 6). Data are expressed as mean ± S.D. B and C, cells (1.2 × 105) were transduced with either the indicated MOI of either Ad.mda-7 or Ad.CMV control virus for 24 h (B) or 150 MOI of either Ad.mda-7 or Ad.CMV control virus for the indicated time (C). Total RNA was extracted and subjected to competitive qRT-PCR analysis of Bcl-x splice variants and the corresponding Bcl-x(L)/(s) mRNA ratios. Data are expressed as mean ± S.D. D and E, The indicated cells (1.2 × 105) were transduced with 150 MOI of either Ad.mda-7 or Ad.CMV control virus. After 24 h, total RNA was extracted and subjected to quantitative/competitive RT-PCR analysis as in panel A. Data are expressed as mean ± S.D. F, the same samples utilized in panel B were subjected to competitive qRT-PCR analysis of Mcl-1 and CD44 splice variants. For all panels, data are representative of six separate determinations on two separate occasions.