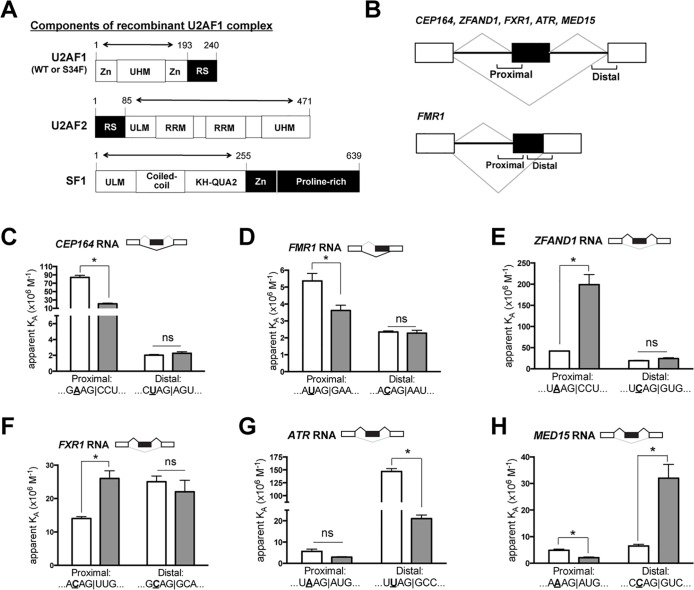
Fig 5. Differential binding of mutant and wild-type U2AF1 complexes to RNA oligonucleotides explains most S34F-associated alterations in RNA splicing.
(A). Cartoons illustrate components of recombinant U2AF1 complexes used in the binding assay. Full-length proteins are shown but only partial sequences (denoted by bi-directional arrows) were used to make recombinant protein complexes. KH-QUA2, K-Homology Quaking motif; RRM, RNA recognition motif domain; RS, arginine/serine-rich domain; UHM, U2AF homology motif; ULM, U2AF ligand motif; ZnF, zinc finger domain. (B). Scheme of alternative splicing patterns for cassette exons (top diagram) and 5' extended exons from competing 3′ splice site selection (bottom). The brackets indicate the positions of RNA oligonucleotides used for the binding assays. Exons are shown as boxes: white boxes indicate invariant exonic sequences and black boxes denote sequences that are incorporated into mRNA (exonic) only when the proximal 3′ splice sites are used. Introns are shown as solid lines. The grey lines represent possible splices. The names of characterized genes that conform to the patterns shown in the upper and lower cartoons are indicated. (C—H). Mutant and wild-type U2AF1 complexes have different affinities (KA’s) for relevant 3′ splice site oligonucleotides. To accomplish the binding assays, the wild-type or mutant U2AF1 protein complexes were titrated into 5' fluorescein-tagged RNA oligonucleotides over a range of concentrations as described in the Supplemental Materials and Methods. RNA sequences from -4 to +3 relative to the 3′ splice sites (vertical lines) in proximal and distal positions are shown. The nucleotide at the -3 position is bolded and underlined. Empty bars, KA for WT U2AF1 complex; grey bar, KA for mutant U2AF1 complex. For ease of comparison between the affinity binding results with S34F-associated splicing, S34F-promoted splices, as determined from RNA-seq data, are shown on top of each bar graph in black lines. The fitted binding curves, full oligonucleotide sequences, and apparent equilibrium dissociation constants are shown in S12 Fig. The relative changes in affinity and use of proximal versus distal splice sites are summarized in S3 Table. The asterisk represents a statistically significant change by unpaired t-tests with Welch’s correction. ns, not statistically significant.