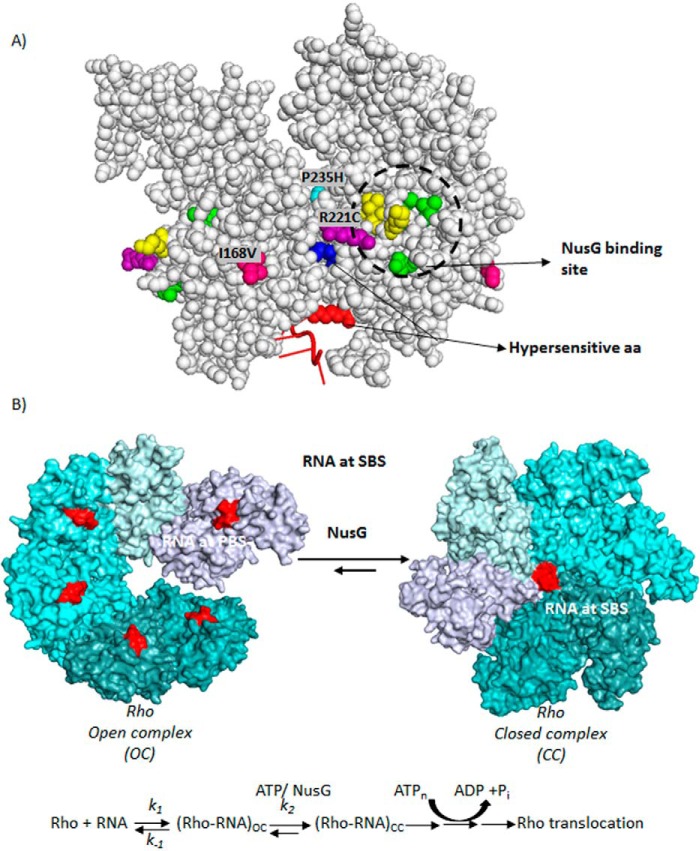
FIGURE 10.
A, locations of the (i) amino acids important for NusG-binding, (ii) Rho* mutants, and (iii) hypersensitive amino acids are indicated on a Rho-dimer structure (PDB code 2HT1). The NusG-binding site is circled, where amino acids in yellow are K224C, V227N, V228N, and those in green are V203L, Q193A, and M205A. B, schematic showing the kinetic/equilibrium steps during the conversion of OC to CC of the Rho hexamer. Putative step(s) those are targeted by NusG are indicated. ATP-binding and hydrolysis steps are also shown. Hexameric structures were based on the coordinates using PDB codes 3ICE and 1PVO, respectively.