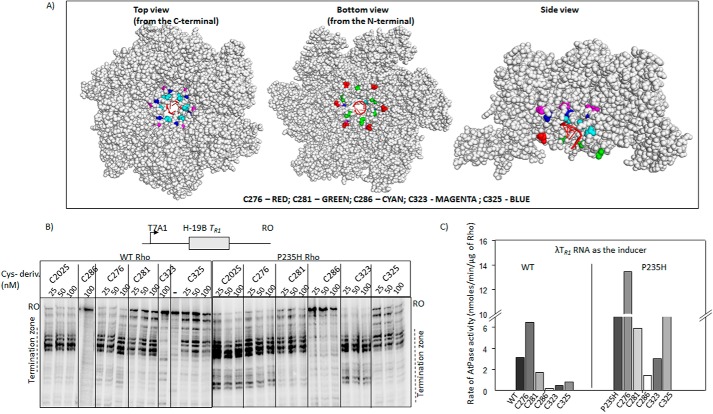
FIGURE 7.
Single Cys derivatives of Rho. A, location of the single cys substitutions on the hexameric structure of Rho (PDB ID 3ICE). The space-filled models were prepared using PyMOL. Positions of the mutations are indicated. The color codes are as follows: T276C, red; S281C, green; T286C, cyan; T323C, magenta; S325C, blue. In first crystal structure the view is from the side of the PBS, in the second is from the side of the SBS, and the third is a side view. The RNA in the central hole of the hexamer is indicated in schematic form as red spirals. B, autoradiograms showing the in vitro Rho-dependent transcription termination assays of the WT and Rho* mutants with single cysteines at different places of the loop structures on the tR1 terminator. The run-off (RO) products and the termination zones of each of the Rho derivatives are indicated. Concentrations of RNAP, DNA, and Rho were 25, 10, and 50 nm, respectively. Template for the in vitro transcription assay was made by PCR amplification on the pRS22 (T7A1-H-19B TR1) using oligo pairs RS83/RK23B. C, bar diagram showing the rates of ATPase activities of all the single cysteine Rho derivatives induced by the λtR1 RNA. Concentrations of Rho and RNA were 25 nm and 20 μm, respectively.