Abstract
Polyadenosine RNA-binding proteins (Pabs) regulate multiple steps in gene expression. This protein family includes the well studied Pabs, PABPN1 and PABPC1, as well as the newly characterized Pab, zinc finger CCCH-type containing protein 14 (ZC3H14). Mutations in ZC3H14 are linked to a form of intellectual disability. To probe the function of ZC3H14, we performed a transcriptome-wide analysis of cells depleted of either ZC3H14 or the control Pab, PABPN1. Depletion of PABPN1 affected ∼17% of expressed transcripts, whereas ZC3H14 affected only ∼1% of expressed transcripts. To assess the function of ZC3H14 in modulating target mRNAs, we selected the gene encoding the ATP synthase F0 subunit C (ATP5G1) transcript. Knockdown of ZC3H14 significantly reduced ATP5G1 steady-state mRNA levels. Consistent with results suggesting that ATP5G1 turnover increases upon depletion of ZC3H14, double knockdown of ZC3H14 and the nonsense-mediated decay factor, UPF1, rescues ATP5G1 transcript levels. Furthermore, fractionation reveals an increase in the amount of ATP5G1 pre-mRNA that reaches the cytoplasm when ZC3H14 is depleted and that ZC3H14 binds to ATP5G1 pre-mRNA in the nucleus. These data support a role for ZC3H14 in ensuring proper nuclear processing and retention of ATP5G1 pre-mRNA. Consistent with the observation that ATP5G1 is a rate-limiting component for ATP synthase activity, knockdown of ZC3H14 decreases cellular ATP levels and causes mitochondrial fragmentation. These data suggest that ZC3H14 modulates pre-mRNA processing of select mRNA transcripts and plays a critical role in regulating cellular energy levels, observations that have broad implications for proper neuronal function.
Keywords: ATP synthase, post-transcriptional regulation, RNA, RNA-binding protein, RNA processing, RNA splicing, MSUT2, Nab2, ZC3H14
Introduction
The fate and function of a cell are determined not only by the spectrum of genes that are expressed but also the spatial and temporal regulation of the resulting mRNAs (1). The processing of mRNAs is referred to as post-transcriptional processing and is critical to ensure proper cell function (2, 3). A myriad of post-transcriptional regulatory RNA-binding proteins execute the post-transcriptional program, which includes events such as the capping, splicing, 3′ end processing, and export of mRNAs (3). These events, which produce a mature export-competent mRNA, are tightly coupled to one another and are subject to extensive quality control to ensure proper processing and to prevent aberrantly processed mRNAs from entering the cytoplasm (4). Therefore, precise expression and function of RNA-binding proteins is critical to ensure the proper gene expression profile of a cell (2). The importance of RNA-binding proteins is illustrated by the number of human diseases that result from mutation or dysregulation of these RNA-binding proteins, including links to cancer (5–7) and neuronal dysfunction (8–11). Understanding the role of newly discovered, or previously uncharacterized, RNA-binding proteins in the post-transcriptional processing of target mRNAs is key in delineating the regulation of gene expression as well as defining molecular mechanisms underlying human disease.
An RNA-binding protein that was recently linked to tissue-specific human disease is the zinc finger polyadenosine RNA-binding protein zinc finger CCCH protein 14 or ZC3H14 (12, 13), which is also termed MSUT2 (14). Mutations in ZC3H14 cause non-syndromic intellectual disability (12), but the mechanism that leads to the disease is unknown. Biochemical studies show that ZC3H14 employs five evolutionarily conserved tandem cysteine3histidine (CCCH) zinc fingers to recognize polyadenosine RNA with high affinity and specificity (15). In contrast, the well studied poly(A)-binding proteins (Pabs),2 poly(A)-binding protein cytoplasmic 1 (PABPC1) and poly(A)-binding protein nuclear 1 (PABPN1), recognize polyadenosine RNA via globular RNA recognition motifs (16–19). Thus ZC3H14 presents a novel mode of polyadenosine RNA recognition and expands the Pab family of RNA-binding proteins (20).
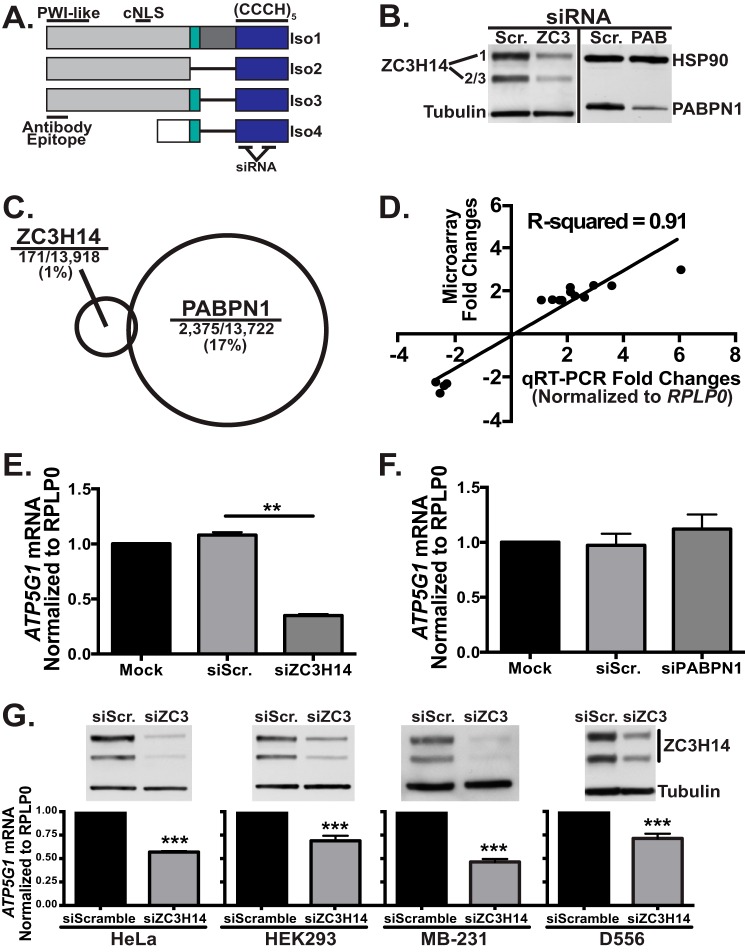
As illustrated in Fig. 1A, ZC3H14 is alternatively spliced to form at least four splice variants that encode four distinct protein isoforms (13). Isoforms 1–3 each contain an N-terminal proline tryptophan isoleucine (PWI)-like domain (13), which mediates interactions with the nuclear pore (21), as well as a predicted classical nuclear localization signal (cNLS) (13). The three longer isoforms of ZC3H14 are localized to the nucleus at steady state (13); however, isoform 4, which lacks the exon containing the predicted cNLS, is localized to the cytoplasm (13). All four described isoforms contain the five tandem C-terminal CCCH zinc fingers and are therefore expected to interact with polyadenosine RNA with similar affinity (13, 15). Patients homozygous for loss-of-function mutations in the ZC3H14 gene have a severe form of autosomal recessive intellectual disability (12), suggesting a critical role for ZC3H14 in mRNA-processing events that are necessary for proper brain function (11).
FIGURE 1.
Knockdown of ZC3H14 decreases ATP5G1 mRNA levels in all cell types examined. A, ZC3H14 is alternatively spliced to form at least four distinct protein isoforms (Iso1–4), three longer isoforms (Iso1–3), and a shorter isoform (Iso4). All isoforms contain the C-terminal (CCCH)5 zinc finger domain (blue) that confers RNA binding. Isoforms 1–3 differ from one another only in selective inclusion of exons 10–12 (teal and dark gray). These isoforms all contain an N-terminal proline tryptophan isoleucine-like (PWI-like) fold as well as a predicted cNLS. Consistent with the presence of a cNLS, Iso1–3 are all localized to the nucleus at steady state. ZC3H14 isoform 4 contains a distinct N-terminal exon (white). As the cNLS is absent from this isoform, Iso4 localizes to the cytoplasm at steady state. The ZC3H14 antibody used in this study recognizes the N-terminal domain of isoforms 1–3 (Antibody Epitope). The siRNAs employed in this study target two independent sequences within the region that encodes for the CCCH zinc fingers. B, to assess knockdown, MCF-7 cells transfected with Scramble (Scr.), ZC3H14 (ZC3), or PABPN1 (PAB) siRNA were subjected to immunoblot analysis with ZC3H14 or PABPN1 antibody and control antibodies to detect tubulin and heat shock protein 90 (HSP90). Robust knockdown of ZC3H14 (∼75–80%) and PABPN1 (∼60–75%) was detected with no effect on tubulin or HSP90 (controls). C, total RNA isolated from MCF-7 cells transfected as in B was used for cDNA generation and hybridization to the Illumina BeadChip microarray platform. A schematic is shown indicating the relative number of transcripts that show a change (>1.5-fold) in steady-state level for each knockdown with size of circle representing fraction of transcripts impacted. Significance analysis of microarrays analysis revealed that 171 out of 13,918 (∼1%) of expressed transcripts in the transfected cells were affected (increased or decreased) by knockdown of ZC3H14 (101 increased and 70 decreased), whereas PABPN1 knockdown modulated 2,375 out of 13,722 (∼17%) expressed transcripts (1,285 increased and 1,090 decreased). D, fold-change values of select affected transcripts identified by the microarray analysis were plotted against fold-changes of the same select transcripts obtained by qRT-PCR analyses. Linear regression was used to determine the R2 value of 0.91, which represents a significant correlation between the results of both analyses and validates the effect on the transcripts analyzed. E and F, total RNA isolated from MCF-7 cells treated with mock transfection (Mock), Scramble siRNA (siScr.), ZC3H14 (siZC3H14, E) or PABPN1 (siPABPN1, F) siRNA was used for cDNA generation and qRT-PCR analysis with transcript-specific primers to detect ATP5G1 and the control RPLP0 mRNA. Knockdown of ZC3H14 (E), but not PABPN1 (F), results in a significant decrease in ATP5G1 steady-state mRNA levels. G, HeLa, HEK293, MB-231, and D556 cells (left to right) were transfected with Scramble or ZC3H14 siRNA. Transfected cells were subjected to immunoblot analysis to confirm knockdown (top) with ZC3H14 and tubulin (control) antibodies as well as qRT-PCR analysis (bottom) with ATP5G1 and RPLP0 (control) primers. Robust knockdown of ZC3H14 in each cell type resulted in a significant decrease in ATP5G1 steady-state mRNA levels. Values represent the mean ± S.E. for n = 3 independent experiments. ** and *** represent p ≤ 0.01 and p ≤ 0.001, respectively.
The RNA recognition motif-containing Pabs, PABPC1 and PABPN1, have well studied and ubiquitous roles in post-transcriptional processing (20); however, the role of ZC3H14 in post-transcriptional mRNA processing is unclear. Extensive work on the Saccharomyces cerevisiae ortholog of ZC3H14, Nab2 (22, 23), reveals that this essential protein plays roles in poly(A) tail length control and mRNA export (24–27), target transcript stability (28), and RNA quality control in the nucleus (29, 30). Genetic analyses performed in Drosophila melanogaster reveal a conserved function of Drosophila Nab2 (dNab2) in post-transcriptional processing, specifically in neurons (31), consistent with the brain dysfunction present in patients. Understanding the role of ZC3H14 in human cells is not only critical to determine the molecular basis for the observed neuronal phenotype in patients but also to integrate the function of ZC3H14 into our current understanding of the Pab family of proteins.
To understand the role of ZC3H14 in human cells, we employed a genome-wide analysis to assess the consequence of ZC3H14 knockdown. Identifying transcripts that are modulated by the loss of ZC3H14 will not only provide useful candidate targets for the analysis of post-transcriptional processing defects but may also provide insight into molecular mechanisms that could contribute to brain dysfunction in patients lacking ZC3H14. In this study, we provide evidence from our genome-wide study that ZC3H14 has specific targets in human cells, as evidenced by the much smaller percentage of transcripts affected by knockdown of ZC3H14 as compared with another nuclear Pab, PABPN1. We selected and identified the ATP5G1 transcript as a specific target of ZC3H14. ATP5G1 encodes one of the proteins that compose the C subunit of ATP synthase (32), which is responsible for the majority of ATP production in eukaryotic cells (33). We demonstrate that knockdown of ZC3H14 in human cells results in a decrease in ATP5G1 steady-state mRNA levels. Our data suggest that ZC3H14 ensures proper pre-mRNA processing of the ATP5G1 transcript prior to transport to the cytoplasm. Finally, depletion of ZC3H14 by siRNA-mediated knockdown causes a decrease in cellular ATP levels and a severe mitochondrial fragmentation phenotype comparable with what is observed upon direct knockdown of ATP5G1. Together, these results support a role for ZC3H14 in modulating specific target mRNAs, including those important for proper mitochondrial function that could be particularly critical in the brain.
Experimental Procedures
Cell Culture
MCF-7 cells (ATCC HTB-22; estrogen receptor-positive breast cancer cell line (34)) were obtained from the ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. DNA plasmids and siRNA (Invitrogen) were transfected into cultured cells using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol.
Plasmids and Chemicals
FLAG fusion constructs were generated using PCR primers that include the FLAG sequence to create N- or C-terminally FLAG-tagged protein isoforms. PCR products were then purified and subcloned into the pcDNA3.1 vector (Invitrogen). To generate the Myc-tagged ZC3H14 constructs, the N-terminal FLAG tag present in the ZC3H14 constructs was removed and replaced (subcloned) with a digested oligonucleotide encoding an N-terminal Myc tag. siRNA-resistant constructs were generated by site-directed mutagenesis (Stratagene) using primers that create silent mutations within the regions targeted by siRNA. For PARP cleavage assays, cells were treated with 1 μm staurosporine (Enzo Life Sciences) for 3 h. Pre-designed Stealth siRNAs (Invitrogen) were employed for knockdown of ZC3H14 and ATP5G1. The ZC3H14 siRNA targeting sequences are as follows: 5′-CACATTCTACCATCCCACCATTAAT-3′ and 5′-TGTTTGTTTGTTCACCCAAATTGTA-3′. The siRNA targeting sequence for ATP5G1 is 5′-TCTCCAGCTCTGATCCGCTGTTGTA-3′. A pre-designed MISSION esiRNA pool (Sigma) was employed for knockdown of UPF1.
Immunoblotting
MCF-7 cells were harvested and washed in 1× PBS and then lysed on ice in RIPA-2 buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0) containing protease inhibitors (PLAC: 3 μg/ml of pepstatin, leupeptin, aprotinin, and chymostatin and 0.5 mm PMSF). Immunoblotting was performed using standard methods (35). Briefly, 30 μg of total protein lysate per sample was resolved by SDS-PAGE and transferred onto a nitrocellulose membrane. For immunoblotting, a 1:5,000 dilution of rabbit polyclonal ZC3H14 (13), rabbit polyclonal PABPN1 (36), or tubulin (Sigma; Clone DM1A) antibody, a 1:2,000 dilution of HSP90 (Santa Cruz Biotechnology; clone F-8) antibody, or a 1:1,000 dilution of UPF1 (Bethyl Laboratories), PARP (Pharmingen; clone 4C10-5), HuR (Santa Cruz Biotechnology; clone 3A2), or Myc (Cell Signaling; clone 9B11) antibody was used followed by 1:3,000 dilutions of HRP-conjugated goat anti-mouse IgG or HRP-conjugated goat anti-rabbit IgG secondary antibodies (Jackson ImmunoResearch).
Microarray Analyses
MCF-7 cells were transfected with siScramble, siZC3H14, or siPABPN1 and collected 48 h later. Transfected MCF-7 cells were lysed using QIAshredder columns (Qiagen), and total RNA was isolated using the RNeasy mini kit (Qiagen). RNA integrity from triplicate samples was assessed on an Agilent Bioanalyzer 2100, and cDNA samples were generated and hybridized to the Illumina HumanHT-12 v4 Expression BeadChip microarray platform by the Emory Integrated Genomics Core. To discover differentially expressed transcripts between treatment groups, a two-class unpaired Significance Analysis of Microarrays analysis was employed with a false discovery rate of 0%. An additional cutoff of 1.5-fold was used to identify significantly affected (increased or decreased) transcripts.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from MCF-7 cells using TRIzol reagent (Invitrogen) in accordance with the manufacturer's instructions. Reverse transcriptase reactions were carried out with Random Primers (hexadeoxynucleotides; Promega) and Moloney murine leukemia virus RT (Invitrogen) using 1 μg of RNA for an estimated concentration of ∼50 ng/μl cDNA per sample that was used for quantitative RT-PCR.
For qRT-PCR analyses, 1 μg of total RNA was transcribed to cDNA as described above. Relative mRNA levels were measured by quantitative PCR analysis of triplicate samples of 5 ng of cDNA with QuantiTect SYBR Green Master Mix using an Applied Biosystems real time machine (ABI). Results were analyzed using the ΔΔCT method (37) and normalized to 18S rRNA or RPLP0 transcript. Statistical significance was determined using either a Student's t test or a one- or two-way analysis of variance. Primer sequences used for qRT-PCR analyses are as follows: 18S rRNA FWD, GAGACTCTGGCATGCTAACTAG, and 18S rRNA REV, GGACATCTAAGGGCATCACAG; RPLP0 FWD, GGGCGACCTGGAAGTCCAACT, and RPLP0 REV, CCCATCAGCACCACAGCCTTC; ATP5G1 FWD, CCTCCTTCTTGAATAGCCCAG, and ATP5G1 REV, CCCCAGCACCAATAAACTTG; ZC3H14 FWD, CTACCATCACCCCATCTCAC, and ZC3H14 REV, AGGGACAATCTGGTTTAGTACAC; Pre-ATP5G1 FWD, GAGTCAGCCACCTGTCCTTATGCC, and Pre-ATP5G1 REV, CTGGTCTGGAACTCCCGTCTGG; Pre-RPLP0 FWD, GTGGCCATGGATCTGCTGGTTGTC, and Pre-RPLP0 REV, CCCACTTTGTCTCCAGTCTTGATCAGCTG; and GAPDH FWD, AAGGTCGGAGTCAACGGATTTGG, and GAPDH REV, GATGACAAGCTTCCCGTTCTC. Pre-validated qRT-PCR primers for ATP5G2, ATP5G3, and the OXPHOS subunit mRNAs NDUFA4, SDHB, UQCRFS1, COXIV, and ATP5B were ordered from SABiosciences. To verify the identity of the PCR products, we resolved the products on an agarose gel (to verify one band/primer set), excised each fragment, and performed TOPO cloning followed by sequencing.
RNA Immunoprecipitation
To probe interactions between ZC3H14 and target mRNAs in whole cell lysates, RNA-IP analyses were performed on MCF-7 cells grown to near confluency in 150-mm plates. Cells were rinsed twice with ice-cold PBS, collected on ice, and resuspended in RNA-IP buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 32 mm NaF, 0.5% Nonidet P-40 in diethyl pyrocarbonate-treated water) supplemented with 1 mm DTT, 100 units/ml RNase OUT (Invitrogen), and 1 cOmplete mini protease inhibitor tablet (Roche Applied Science; 1 tablet/10 ml of buffer). Cells were sonicated on ice five times (program, 9 s at 0.5 output) and then passed through a 27-gauge syringe five times and placed back on ice for 20 min with occasional vortexing. Lysates were spun at 13,000 rpm for 10 min at 4 °C, and protein concentration was determined with a standard BCA assay. Protein G magnetic beads (DYNA beads; Invitrogen) were rinsed and resuspended in RNA-IP buffer and incubated with pre-immune rabbit serum or an equal volume of N-terminal ZC3H14 antibody (13) for 1 h at room temperature. Bead/antibody and bead/pre-immune samples were rinsed in RNA-IP buffer and added to clarified cell lysates, followed by incubation at 4 °C overnight while tumbling end over end (10% removed prior to overnight incubation for input samples). After incubation, beads were magnetized; unbound samples were collected (10% of input) and washed five times with ice-cold RNA-IP buffer. ZC3H14/RNA complexes were eluted and isolated with TRIzol reagent (Invitrogen) and purified according to the manufacturer's instructions.
To determine interactions between ZC3H14 and target RNAs in separate cellular compartments, RNA-IP assays were carried out as described above, with the following alterations. Confluent MCF-7 cells were rinsed twice with ice-cold PBS, collected on ice, and subjected to nucleocytoplasmic fractionation (described below). Upon isolation and separation of the cytoplasmic lysates, the nuclear fraction was rinsed once with ice-cold TSE buffer (10 mm Tris, 300 mm sucrose, 1 mm EDTA, 0.1% Nonidet P-40, pH 7.5), sonicated, and passed through a syringe as described above. Nuclear samples were incubated on ice for 20 min with occasional vortexing, followed by a 10-min spin at 13,000 rpm at 4 °C. The protein concentration from each compartment was determined by a standard BCA assay. Lysates from each compartment were subjected to immunoprecipitation as described above.
RNA isolated from input and bound fractions for pre-immune and ZC3H14 samples were subjected to RNA isolation and qRT-PCR as described above. mRNA levels in the ZC3H14-bound fractions (for whole cell and within each compartment) were normalized to input levels and then compared by fold-enrichment over pre-immune control.
mRNA Stability Assays
To measure mRNA stability in MCF-7 cells, 5 μg/ml actinomycin D (Sigma) was added to the growth medium to inhibit transcription, and cells were harvested 0, 6, 9, and 12 h later. To measure mRNA stability in MCF-7 cells with reduced ZC3H14 levels, cells were transfected with ZC3H14 siRNA 24 h after seeding. Following a 48-h incubation with siRNA, 5 μg/ml actinomycin D was added to the growth medium, and cells were harvested 0, 6, 9, and 12 h later. Total RNA was extracted from cells (described above) and analyzed by qRT-PCR. Half-lives were determined by normalization to 18S rRNA and to time 0.
Nucleocytoplasmic Fractionation
To isolate distinct pools of RNA and protein from nuclear and cytoplasmic fractions, MCF-7 cells were collected on ice, spun down, and resuspended in ice-cold fractionation buffer (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 3 mm MgCl2, 0.5% (v/v) Nonidet P-40) supplemented with 1 mm DTT, 100 units/ml RNase OUT (Invitrogen), and 1 mini cOmplete protease inhibitor tablet (Roche Applied Science; 1 tablet/10 ml of buffer) for 10 min on ice. Cell lysates were centrifuged at 1,000 × g for 5 min, and the supernatant or cytoplasmic fraction was separated from the nuclear pellet. RNA was isolated from each fraction with TRIzol reagent (Invitrogen), and protein samples were prepared with RIPA-2 buffer (as described above for immunoblotting).
Cellular ATP Assays
Cells were plated in 6-well dishes and treated for 48 h with siScramble, siZC3H14, or siATP5G1 siRNA (Invitrogen) according to the manufacturer's protocol. Separate sets of cells were treated with vehicle control (DMSO; Sigma) or 1 μm rotenone (Enzo Life Sciences) for 3 h. All cells were harvested, counted side-by-side, and then lysed using a one-step boiling water cellular ATP extraction as described previously (38). Total cellular ATP levels were assayed with the Enliten ATP Assay System (Promega) and normalized to siScramble or vehicle control.
Immunofluorescence and Imaging
MCF-7 cells were plated and transfected with Scramble or ZC3H14 siRNA (Invitrogen) using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol. Following a 48-h transfection, cells were re-plated at a 1:2 dilution onto Matrigel (BD Biosciences)-coated coverslips and allowed to attach overnight. The following day, cells were fixed in 4% paraformaldehyde in PBS (15 min at room temperature) and then permeabilized in Triton X-100 (0.1% in PBS, 10 min), blocked (1% BSA in PBS, 1 h), and stained overnight with a 1:1,000 dilution of cytochrome c (BD Biosciences; clone 6H2.B4) or a 1:5,000 dilution of HSP60 (Enzo Life Sciences; clone LK-2) antibodies diluted in blocking buffer. The coverslips were then washed four times with PBS (5 min) and stained with a 1:500 dilution of anti-mouse 546 secondary antibody (Alexa fluorophores, Invitrogen). Coverslips were washed four times, stained with Hoechst 33342, and mounted using Prolong Antifade (Invitrogen). Representative cells were imaged using an Olympus FluoView 1000 confocal microscope using a ×100 objective (NA 1.45) with laser excitation at 543 nm.
For quantitative analyses, a cell was scored as “normal” if its mitochondria appeared predominantly tubular, and “fragmented” if mitochondria appeared predominantly small and spherical. For each experiment at least 100 cells were scored. Values are represented as mean ± S.E. from three independent experiments. Statistical analysis was performed using a one-way analysis of variance, and a value of p ≤ 0.05 was considered significant.
Results
Knockdown of ZC3H14 Decreases ATP5G1 mRNA Levels
Previous studies of ZC3H14 orthologs in model organisms suggest a spectrum of post-transcriptional functions (24, 25, 28) that may influence the fate of bound target transcripts (29). To examine the function of ZC3H14 in human cells, we sought to identify candidate target mRNA transcripts via an unbiased genome-wide approach. To accomplish this goal, we performed a microarray analysis on MCF-7 cells, a commonly used human breast cancer cell line (ATCC HTB-22), transfected with either control Scramble siRNA or siRNA targeting ZC3H14. To compare ZC3H14 to a more conventional Pab, we also performed knockdown of the well studied nuclear Pab, PABPN1 (20, 39, 40). PABPN1 is predicted to bind to and regulate the 3′ end processing of numerous polyadenylated mRNAs (41–43) and has the potential to regulate the steady-state level of many cellular mRNAs. A recent study demonstrated that knockdown of PABPN1 in HeLa cells resulted in a ≥1.5-fold change of ∼13.5% of expressed transcripts in those cells (3 and 10.5% increased and decreased, respectively) (44). If ZC3H14, as another nuclear Pab, also binds and regulates a large number of polyadenylated transcripts, we would expect to see a similar number of mRNA transcripts affected upon depletion of each Pab.
As shown in Fig. 1B, we achieve robust knockdown of ZC3H14 and PABPN1, as determined by immunoblotting with a ZC3H14 antibody that detects the nuclear isoforms of ZC3H14 (Fig. 1A) (13) or a PABPN1 antibody (36). Knockdown experiments were performed under conditions where no change in cell growth or viability was detected to minimize indirect effects. To identify differentially expressed transcripts between control cells and cells depleted of ZC3H14 or PABPN1, total RNA was used for cDNA generation and hybridized to the Illumina HumanHT-12 v4 Expression BeadChip microarray platform. Significance analysis of Microarrays was employed to determine statistically significant differentially expressed genes (≥1.5-fold change) between treatment groups. As shown in Fig. 1C, knockdown of PABPN1 affected the steady-state level of 2,375 or ∼17% of transcripts expressed in these cells, as calculated from our analyses (1,285 increased and 1,090 decreased transcripts out of 13,722 total). However, knockdown of ZC3H14 resulted in a change in steady-state level of only 171 or ∼1% of expressed transcripts (101 increased and 70 decreased transcripts out of 13,918 total). To validate the results of the microarray, we performed qRT-PCR analyses with primers to a selection of the ZC3H14-affected transcripts (Fig. 1D) normalized to the housekeeping gene, RPLP0, which was unaffected by ZC3H14 or PABPN1 knockdown. As shown in Fig. 1D, the fold-changes calculated by qRT-PCR analysis correlate strongly (r2 = 0.91) with the results of the microarray, supporting the validity of the genome-wide study.
The observation that PABPN1 knockdown affects a significant percentage of expressed transcripts supports the well studied role of PABPN1 in 3′ end processing and polyadenylation of numerous mRNA transcripts (41–43) and is consistent with the previous study performed in HeLa cells (44). We hypothesize that the significantly smaller number of transcripts that show altered steady-state level upon ZC3H14 knockdown represents a pool of transcripts that are regulated by ZC3H14 in a specific manner.
To determine how ZC3H14 alters target transcripts, we selected one robustly affected target for further analysis. The ATP synthase subunit C1, or ATP5G1, transcript is robustly decreased (∼3-fold) upon knockdown of ZC3H14 (Fig. 1E). In contrast to ZC3H14 knockdown, ATP5G1 mRNA levels are not affected by knockdown of PABPN1 (Fig. 1F), suggesting that it is not a general target of regulation by nuclear Pabs and that ATP5G1 may in fact be specifically regulated by ZC3H14.
To confirm that the effect of ZC3H14 on ATP5G1 steady-state levels is not limited to MCF-7 cells, we extended our analysis to a number of different human cells lines. As shown in Fig. 1G, we achieved robust knockdown of ZC3H14 in HeLa (cervical adenocarcinoma, ATCC CCL-2), HEK293 (embryonic kidney, ATCC CRL-1573), MB-231 (breast adenocarcinoma, ATCC HTB-26), and D556 (medulloblastoma (45)) cells. Total RNA isolated from these cells was used for qRT-PCR analysis with ATP5G1 and the control ribosomal protein, large P0 (RPLP0) primers. Results of this analysis revealed a consistent reduction of ATP5G1 steady-state mRNA levels across all cell lines tested.
A previous study demonstrated that MCF-7 cells produce 80% of their ATP by oxidative phosphorylation (the remaining 20% coming from glycolysis) (46), whereas HeLa cells and many other cancer cell lines primarily use glycolysis for ATP production (47). Because we eventually wanted to investigate whether knockdown of ZC3H14 had an impact on cellular ATP levels and energy metabolism via regulation of ATP5G1, we used the MCF-7 cell line for our analyses throughout this study.
Re-expression of ZC3H14 Isoform 1 (Iso1) Partially Rescues the Decrease in ATP5G1 Transcript
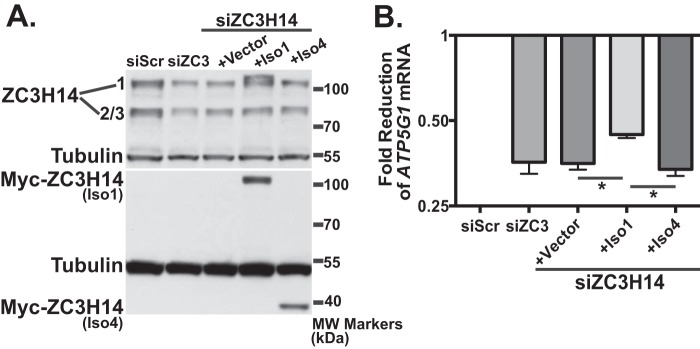
To confirm that the effect on ATP5G1 is due specifically to depletion of ZC3H14, we validated our results with two independent siRNAs and also performed a rescue experiment with siRNA-resistant Myc-tagged ZC3H14 constructs. We achieve robust knockdown of ZC3H14, accompanied by a decrease in ATP5G1 mRNA, with each individual ZC3H14 siRNA (supplemental Fig. 1). For the rescue experiment, we assessed the role of nuclear and cytoplasmic isoforms of ZC3H14. We co-transfected MCF-7 cells with ZC3H14 siRNA together with plasmids expressing Myc-ZC3H14 Iso1 (nuclear; see Fig. 1A) or Myc-ZC3H14 Iso4 (cytoplasmic; see Fig. 1A). As shown in Fig. 2A, both Myc-ZC3H14 isoform 1 and isoform 4 are robustly expressed. RNA isolated from these cells was used for cDNA generation and subsequent qRT-PCR analysis with primers to detect ATP5G1 and a control transcript, 18s rRNA. As shown in Fig. 2B, expression of the Myc-tagged ZC3H14 isoform 1 significantly rescues steady-state ATP5G1 mRNA levels when compared with knockdown alone. In contrast, Myc-ZC3H14 isoform 4 does not rescue the effect of ZC3H14 knockdown on ATP5G1 levels, suggesting that the nuclear isoform of ZC3H14 is required to regulate ATP5G1 mRNA. Although the rescue achieved with Myc-ZC3H14 isoform 1 is significant, levels of ATP5G1 mRNA are not fully restored. This partial rescue could be due to expression of a single isoform of ZC3H14 (Iso1) when the endogenous gene encodes multiple isoforms of the protein (see Fig. 1A). Together, these results demonstrate that the decrease of ATP5G1 mRNA levels is due to depletion of nuclear ZC3H14.
FIGURE 2.
Re-expression of ZC3H14 isoform 1 restores ATP5G1 transcript levels. To rescue the effect of ZC3H14 knockdown on ATP5G1 mRNA levels, MCF-7 cells were transfected with either Scramble (siScr.) or ZC3H14 (siZC3) siRNA alone or co-transfected with ZC3H14 siRNA and pcDNA3 (Vector), Myc-tagged ZC3H14 isoform 1 (Iso1), or Myc-tagged ZC3H14 isoform 4 (Iso4) for 48 h. The Myc-tagged ZC3H14 constructs harbor silent mutations in the ZC3H14 siRNA-targeting regions and are therefore refractory to siRNA knockdown. Transfected cells were subjected to immunoblot analysis (A) with ZC3H14, tubulin (control), or Myc antibody and qRT-PCR analysis (B) with primers specific to ATP5G1 and control 18s rRNA. Scramble control values are set to 1.0, and fold-reduction of ATP5G1 mRNA is represented on a log2 axis. A significant rescue of ATP5G1 mRNA upon re-expression of Myc-Iso1 but not -Iso4 is indicated by *, which represents p ≤ 0.05.Values represent the mean ± S.E. for n = 3.
ZC3H14 Specifically Regulates the ATP5G1 Transcript
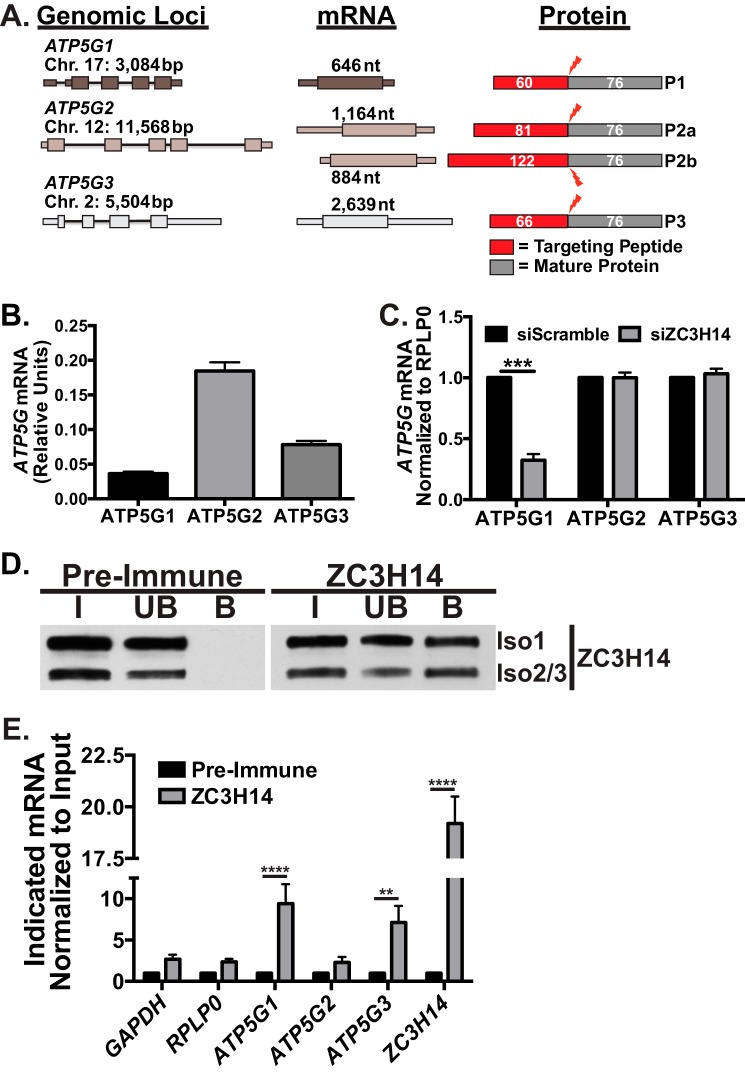
The ATP5G1 transcript encodes one of the 15 subunits of ATP synthase (33), specifically subunit C. Subunit C is one of the transmembrane components of the F0 portion of ATP synthase, which is responsible for proton translocation across the inner mitochondrial membrane (33). The proton-driven rotation of subunit C and subsequent rotation of the F1 stalk provide energy for ATP synthesis by subunit α and β in the mitochondrial matrix. Interestingly, the C subunit is composed of multiple copies (eight in humans (48)) of the C protein, which are derived from multiple genomic loci (32, 49, 50). As schematized in Fig. 3A, the ATP5G1, ATP5G2, and ATP5G3 genes are nuclear-encoded genes that are transcribed from three separate genomic loci and give rise to unique mature mRNAs. These unique mRNAs encode precursor proteins that ultimately produce identical mature proteins that initially include distinct N-terminal peptides that are cleaved upon mitochondrial import (32, 49, 50). Upon cleavage of the mitochondrial targeting peptides, the identical protein products derived from different genes are integrated into the C subunit ring at an undetermined stoichiometry.
FIGURE 3.
ZC3H14 specifically regulates the ATP5G1 transcript. A, ATP5G1, ATP5G2, and ATP5G3 transcripts are transcribed from three separate genomic loci on chromosomes (Chr.) 17, 12, and 2, respectively. The ATP5G1, -2, and -3 genes and their corresponding mRNAs have varying lengths (reported in base pairs (bp) or nucleotides (nt); thin bars = introns, thicker bands = UTRs, and boxes = coding regions) and encode distinct protein products (P1, P2, and P3). ATP5G2 is alternatively spliced to form two distinct mRNAs and subsequent protein products, P2a and P2b. The encoded protein products contain identical C termini with variable N-terminal mitochondrial targeting peptides (red, with the number of amino acids indicated) that are cleaved (red lightning bolt) upon import into the mitochondria. The resulting mature protein products (dark gray) are completely identical in amino acid sequence (76 amino acids). B, total RNA isolated from MCF-7 cells was used for qRT-PCR analysis with primers specific to each of the ATP5G mRNAs. The relative value of each ATP5G mRNA was calculated by 2∧−Ct and is reported as relative units. C, MCF-7 cells transfected with Scramble control or ZC3H14 siRNA were subjected to RNA isolation and qRT-PCR analysis with primers specific to all three ATP5G mRNAs as well as the control transcript, RPLP0. Values are set to 1.0 for siScramble and normalized to RPLP0. Knockdown of ZC3H14 results in a specific and robust decrease in ATP5G1 steady-state mRNA levels. D, endogenous nuclear isoforms of ZC3H14 were immunoprecipitated from MCF-7 cells using either ZC3H14 antibody-bound protein A/G beads or control rabbit pre-immune serum-coated beads. Proteins from the input (I), unbound (UB), and bound (B) fractions were resolved on an SDS-polyacrylamide gel and subjected to immunoblotting with ZC3H14 antibody. The nuclear ZC3H14 isoforms were detected in the ZC3H14-bound fraction but not the pre-immune bound fraction. E, RNA isolated from the ZC3H14 RNA-IP was subjected to qRT-PCR analyses with GAPDH, RPLP0, ATP5G1, ATP5G2, ATP5G3, and ZC3H14 primers. mRNA levels in the ZC3H14 bound fractions were normalized to input levels and then compared by fold-enrichment over pre-immune control. Significant enrichment of ATP5G1, ATP5G3, and ZC3H14 transcripts was observed with ZC3H14 IP. Values represent the mean ± S.E. for n = 3 independent experiments. **, ***, and **** represent p ≤ 0.01, p ≤ 0.001, and p ≤ 0.0001, respectively.
Recent work suggests that although the three subunit C proteins are identical in sequence, they cannot functionally substitute for one another and are all required to constitute a fully functional C subunit (51, 52). Previous studies show that the ATP5G1 transcript is expressed at the lowest levels among the three transcripts and is regulated distinctly from ATP5G2 or ATP5G3 (53, 54). To determine the relative levels of ATP5G1, -2, and -3 in MCF-7 cells, we isolated RNA from untreated cells and performed qRT-PCR analysis with transcript-specific primers. As shown in Fig. 3B, the steady-state level of ATP5G1 mRNA is significantly lower than the level of ATP5G2 or ATP5G3 in these cells, consistent with previous observations (53, 54). To determine whether the effect of ZC3H14 knockdown is specific to ATP5G1, we knocked down ZC3H14 and analyzed the steady-state level of each ATP5G mRNA in these samples. Consistent with the results of the microarray, ATP5G1 mRNA was the only subunit C mRNA affected by ZC3H14 knockdown, suggesting that ZC3H14 specifically regulates expression of ATP5G1 (Fig. 3C).
ZC3H14 Binds to mRNAs
Extensive in vitro binding studies demonstrate that ZC3H14 recognizes polyadenosine RNA via five evolutionarily conserved CCCH zinc fingers (15), suggesting that ZC3H14 could bind to any and all polyadenylated mRNAs. However, in our microarray analysis, we observed a small number of transcripts that showed significant changes in steady-state levels upon ZC3H14 knockdown. We hypothesized that this relatively small number of affected transcripts represents a set of target transcripts that are both bound and regulated at the steady-state RNA level by ZC3H14. To test whether ZC3H14 binds to ATP synthase subunit C mRNAs, we subjected MCF-7 cells to RNA-IP analyses using ZC3H14 antibody-conjugated protein A/G beads. As shown in Fig. 3D, ZC3H14 is enriched in the anti- ZC3H14 bound fraction with no purification in the pre-immune control bound fraction. To identify RNA transcripts that co-purify with ZC3H14, qRT-PCR analysis was performed (Fig. 3E). ATP5G1 mRNA is significantly enriched with ZC3H14. Significant enrichment of ATP5G3 mRNA is also detected, suggesting that although ZC3H14 can bind to both transcripts, it specifically modulates the steady-state level of only ATP5G1 (Fig. 3C).
A previous study in S. cerevisiae demonstrated that the ZC3H14 ortholog, Nab2, binds and autoregulates the levels of its own mRNA transcript via an internal encoded stretch of 26 adenosines within the 3′UTR (55). Like Nab2, the human ZC3H14 3′UTR contains an internal stretch of 15 adenosines that could confer binding by ZC3H14. Consistent with many other RNA-binding proteins that bind their own transcripts (56), we observe robust enrichment of ZC3H14 mRNA upon purification of ZC3H14, suggesting that this mode of autoregulation is conserved in human cells. A number of other transcripts, including GAPDH, RPLP0, and ATP5G2, were not significantly enriched upon ZC3H14 purification. Together, these results suggest that ZC3H14 binds to specific transcripts, such as ATP5G1, in MCF-7 cells.
ZC3H14 Modulates the Decay of ATP5G1 mRNA
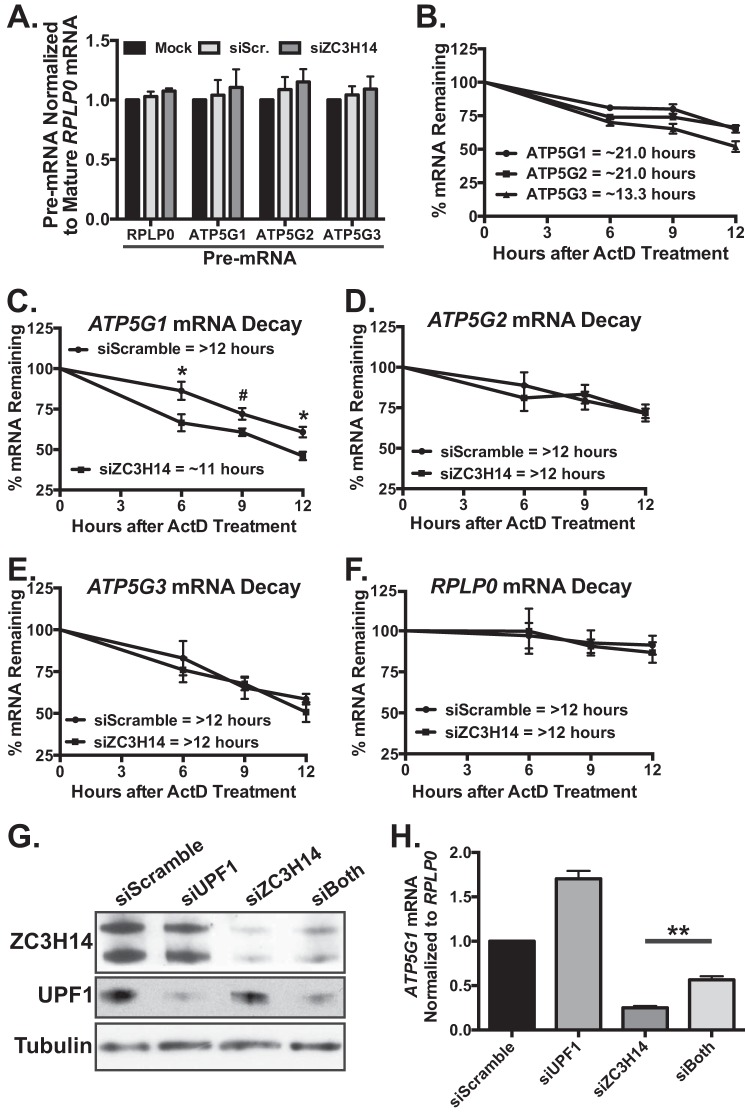
The finding that ZC3H14 knockdown decreases steady-state ATP5G1 mRNA levels suggests that ZC3H14 modulates the transcription, processing, and/or the stability of the ATP5G1 transcript. Previous work in orthologous systems demonstrates that ZC3H14 has specificity for polyadenosine RNA (15) and plays a role in poly(A) tail length control (24), mRNA export (25), target transcript stability (28), and RNA quality control in the nucleus (29, 30); however, we cannot rule out the possibility that ZC3H14 affects ATP5G1 transcription. To test for transcriptional changes to ATP5G1, -2, or -3 upon knockdown of ZC3H14, we performed qRT-PCR analyses with transcript-specific primers to amplify the pre-mRNAs for each of the three subunit transcripts (see Fig. 6B). As shown in Fig. 4A, no change in steady-state pre-mRNA levels of ATP5G1, ATP5G2, or ATP5G3 is detectable following treatment with control or ZC3H14 siRNA, suggesting that a decrease in ZC3H14 does not alter transcription of the ATP5G mRNAs. As expected, siRNA treatment did not impact the steady-state level of the negative control transcript, RPLP0, pre-mRNA.
FIGURE 6.
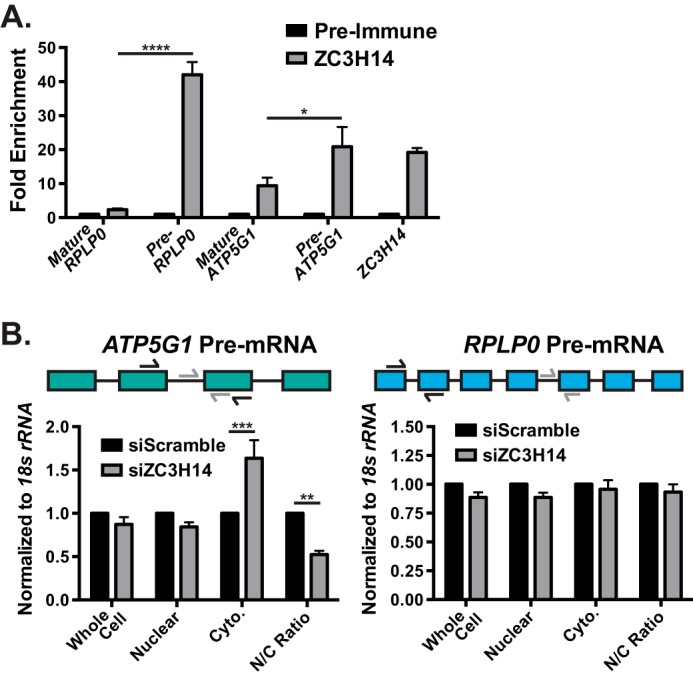
ZC3H14 interacts with ATP5G1 pre-mRNA. A, to determine whether ZC3H14 interacts with ATP5G1 pre-mRNA, RNA isolated from ZC3H14-precipitated MCF-7 cell lysates (see Fig. 3D) were subjected to qRT-PCR analysis with mature RPLP0 mRNA, RPLP0 pre-mRNA (Pre-RPLP0), mature ATP5G1 mRNA, ATP5G1 pre-mRNA (Pre-ATP5G1), and ZC3H14 primers. Consistent with the results from Fig. 3E, we observe significant enrichment of ATP5G1 and ZC3H14 mRNAs in the ZC3H14 bound fraction. Interestingly, we observe significantly higher enrichment of RPLP0 and ATP5G1 pre-mRNA levels compared with their respective mature transcripts. B, total RNA isolated from cells transfected with Scramble and ZC3H14 siRNA followed by nucleocytoplasmic fractionation (as described under “Experimental Procedures”) were subjected to qRT-PCR analysis with primers to detect ATP5G1 (left) and RPLP0 (right) pre-mRNA. A schematic of the coding exons (colored boxes) and introns (black line) of the ATP5G1 and RPLP0 transcripts are represented above the respective graph. Primers used to detect pre-mRNA and mature mRNA are represented with gray and black arrows, respectively. The nuclear/cytoplasmic (N/C) ratio of ATP5G1 pre-mRNA is significantly decreased upon ZC3H14 knockdown, likely due to the increased cytoplasmic levels of ATP5G1 pre-mRNA. The nuclear/cytoplasmic ratio of RPLP0 pre-mRNA is unchanged upon ZC3H14 knockdown. Data points represent the mean ± S.E. for n = 3 independent experiments. *, **, ***, and **** represent p ≤ 0.05, p ≤ 0.01, p ≤ 0.001, and p ≤ 0.0001, respectively.
FIGURE 4.
ZC3H14 modulates the decay of ATP5G1 mRNA. A, total RNA isolated from MCF-7 cells treated with mock transfection, Scramble, or ZC3H14 siRNA was used for qRT-PCR analysis with primers that amplify ATP5G1, -2, -3, and RPLP0 pre-mRNAs as well as mature RPLP0 mRNA. Pre-mRNA levels of each transcript are normalized to levels of mature RPLP0 (set to 1.0), and no significant difference was observed in pre-mRNA levels of these transcripts upon ZC3H14 knockdown. B, MCF-7 cells were treated with the transcriptional inhibitor, ActD, and collected at the indicated time points after drug addition. Total RNA isolated from ActD-treated cells was subjected to qRT-PCR analysis with ATP5G1, ATP5G2, ATP5G3, and 18S rRNA (control) primers. mRNA levels were normalized to time 0 and are represented as % of amount present at time 0. The predicted half-lives of ATP5G1 and ATP5G2 were calculated to be ∼21.0 h, although the predicted half-life ATP5G3 was shorter at ∼13.3 h. To examine differences in stability of the ATP5G transcripts upon ZC3H14 knockdown, MCF-7 cells transfected with Scramble or ZC3H14 siRNA were treated with ActD and collected at the indicated time points after drug addition. qRT-PCR analysis of total RNA isolated from these samples with primers specific to ATP5G1, -2, -3, and RPLP0 mRNAs demonstrates a modest but significant decrease in ATP5G1 mRNA stability (C) with no difference in the decay rate of the other transcripts examined (D–F). mRNA levels were normalized to time 0 and are represented as % mRNA remaining. To determine whether ZC3H14 is involved in proper processing of ATP5G1 mRNA, MCF-7 cells were transiently transfected with Scramble, UPF1, ZC3H14, or UPF1/ZC3H14 (siBoth) siRNA. Transfected cells were subjected to immunoblot analysis (G) with ZC3H14, UPF1, and tubulin (control) antibodies and qRT-PCR analysis (H) with primers specific to ATP5G1 and control transcript, RPLP0. Double knockdown of ZC3H14 and UPF1 results in a significant rescue of ATP5G1 mRNA levels compared with ZC3H14 knockdown alone, suggesting that loss of ZC3H14 results in a pre-mRNA processing defect of ATP5G1 upstream of NMD. Data points represent the mean ± S.E. for n = 3 independent experiments. *, **, and # represent p ≤ 0.05, p ≤ 0.01, and p = 0.056, respectively.
The decrease in ATP5G1 mRNA levels upon ZC3H14 knockdown without any identifiable change in ATP5G1 transcription suggests that ZC3H14 may modulate the stability of the ATP5G1 transcript. To assess any changes in the half-life of ATP5G1 upon ZC3H14 knockdown, we first determined the half-life of the ATP5G transcripts in MCF-7 cells. To do this, we treated cells with the potent transcriptional inhibitor, actinomycin-D (ActD) (57), and collected cells at 0, 6, 9, and 12 h after drug addition. To calculate the percent mRNA remaining over the time course, we performed qRT-PCR analysis with primers specific to the ATP5G transcripts as well as the control RPLP0 transcript (Fig. 4B). As shown in Fig. 4B, the ATP5G transcripts show a relatively slow decay over the course of the experiment, with predicted half-lives of more than 12 h, which is longer than the average mRNA half-life of ∼9 h (58). These results suggest that the ATP5G mRNAs are fairly stable transcripts.
To determine whether ZC3H14 modulates the stability of the ATP5G mRNAs, we transfected MCF-7 cells with scrambled and ZC3H14-specific siRNA followed by treatment with ActD. Cells collected at 0, 6, 9, and 12 h after ActD treatment were analyzed by qRT-PCR to assess % ATP5G1 mRNA remaining relative to time 0. We achieve robust knockdown of ZC3H14 in these samples, and the steady-state level of ZC3H14 remains low in the siZC3H14 samples over the time course of the experiment (supplemental Fig. 2A). As shown in Fig. 4C, qRT-PCR analysis of the transfected cell populations reveals a small but significant decrease in ATP5G1 mRNA stability upon knockdown of ZC3H14 as compared with control Scramble siRNA. The predicted half-life of ATP5G1 mRNA from the siScramble samples is 17 h, compared with an 11-h half-life upon ZC3H14 knockdown. We do not, however, observe a significant difference in the decay of the ATP5G2 and -3 transcripts (Fig. 4, D and E) or the control transcript, RPLP0 (Fig. 4F), providing further evidence for specific regulation of the ATP5G1 transcript by ZC3H14. These results suggest that one possible mode of ZC3H14-mediated regulation of ATP5G1 is through altered transcript stability. However, transcript decay typically occurs in the cytoplasm (59), and we observe rescue of ATP5G1 mRNA levels specifically with a nuclear isoform of ZC3H14.
Improperly processed transcripts that escape to the cytoplasm can be rapidly degraded via the nonsense-mediated decay (NMD) machinery (60, 61). One of the primary effectors of NMD is the UPF1 protein, which cooperates with other NMD components to recruit various mRNA decay factors (60–62). We hypothesize that the observed decrease in ATP5G1 steady-state mRNA levels upon knockdown of ZC3H14 could be due to aberrant export of improperly processed ATP5G1 transcripts to the cytoplasm resulting in rapid degradation via NMD. To test this hypothesis, we analyzed ATP5G1 and RPLP0 transcript levels from cells treated with siRNA targeting UPF1, ZC3H14, or both. As shown in Fig. 4G, we achieve robust knockdown of both ZC3H14 and UPF1 in their respective samples. Knockdown of UPF1 causes an increase in the steady-state level of the ATP5G1 transcript (Fig. 4H, siUPF1) suggesting that ATP5G1 can be regulated by NMD. Consistent with this suggestion, double knockdown of ZC3H14 and UPF1 results in a significant rescue of ATP5G1 mRNA levels compared with knockdown of ZC3H14 alone (Fig. 4H), suggesting that knockdown of ZC3H14 disrupts a pre-mRNA-processing event upstream of cytoplasmic NMD.
ZC3H14 Interacts with ATP5G1 mRNA in the Nucleus
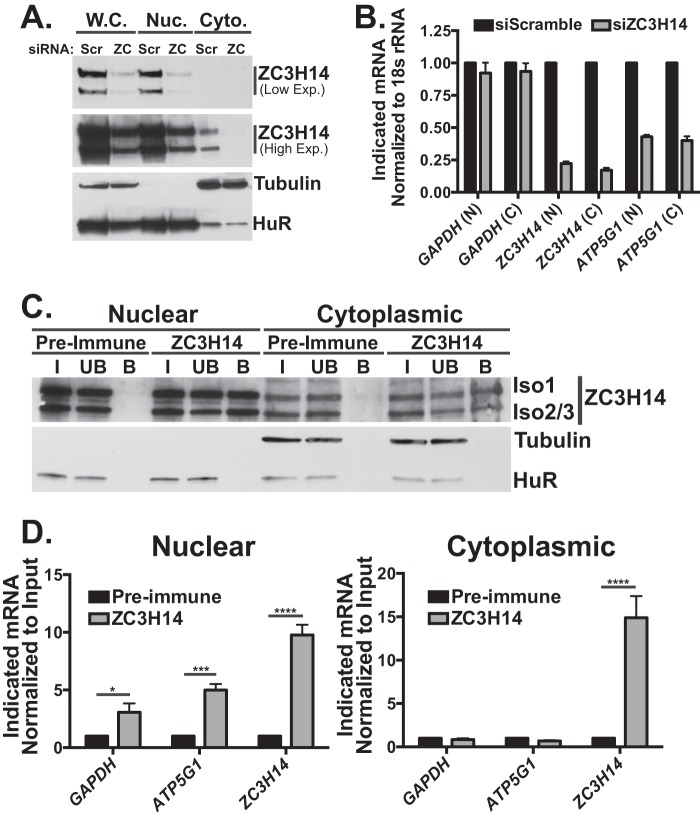
Fig. 2 provides evidence that nuclear ZC3H14 (isoform 1) partially rescues ATP5G1 transcript levels, suggesting a nuclear role for ZC3H14 in regulating ATP5G1. To extend this result and examine the effect of ZC3H14 knockdown in the nuclear and cytoplasmic compartments, we transfected cells with Scramble or ZC3H14 siRNA and collected cells 48 h later. Whole cell, nuclear, and cytoplasmic lysates were collected by nucleocytoplasmic fractionation, as described under “Experimental Procedures.” To confirm robust knockdown in these samples as well as efficient fractionation, we immunoblotted with ZC3H14, Tubulin, and HuR antibodies (Fig. 5A). As expected, tubulin is enriched in the cytoplasmic fraction, and the RNA-binding protein, HuR (63), is detected primarily in the nuclear fraction, confirming efficient fractionation of these cells. The lower exposure (Fig. 5A, Low Exp.) blot of ZC3H14 demonstrates robust knockdown of ZC3H14 as well as clear steady-state nuclear localization of ZC3H14. The higher exposure (High Exp.) blot reveals a small pool of ZC3H14 that is either normally present in the cytoplasm of these cells or could be the result of inefficient fractionation; however, this observed fraction of ZC3H14 is also reduced upon ZC3H14 knockdown (Fig. 5A).
FIGURE 5.
ZC3H14 binds to ATP5G1 mRNA in the nucleus. To determine whether ZC3H14 knockdown impacts ATP5G1 mRNA levels in a specific compartment, MCF-7 cells were transiently transfected with either Scramble (Scr.) or ZC3H14 (ZC) siRNA and then fractionated to analyze the nuclear and cytoplasmic compartments. A, protein from whole cell (W.C.), nuclear (Nuc.), and cytoplasmic (Cyto.) samples were subjected to immunoblot analysis with ZC3H14, tubulin (cytoplasmic), and HuR (nuclear) antibodies. As expected and consistent with efficient fractionation, HuR and tubulin display primarily nuclear and cytoplasmic localizations, respectively. Consistent with a previous study demonstrating steady-state nuclear localization of ZC3H14 in HeLa cells (13), we detect ZC3H14 primarily in the nucleus of MCF-7 cells. Robust knockdown of ZC3H14 in the whole cell and nuclear fractions is shown in the lower exposure blot (Low Exp.). A higher exposure (High Exp.) of the same blot demonstrates robust knockdown of the small cytoplasmic pool of ZC3H14 in the cytoplasmic fraction as well. B, total RNA isolated from samples in A was used for cDNA generation and qRT-PCR analysis with GAPDH, ZC3H14, and ATP5G1 primers. Knockdown of ZC3H14 resulted in a robust decrease of ZC3H14 and ATP5G1 steady-state mRNA levels detected in both the nucleus and cytoplasm. To determine whether ZC3H14 interacts with ATP5G1 mRNA in the nucleus and/or cytoplasm, MCF-7 cells were subjected to nucleocytoplasmic fractionation followed by RNA-IP, as described under “Experimental Procedures.” C, proteins from the input (I), unbound (UB), and bound (B) fractions were subjected to immunoblot analysis with ZC3H14 antibody as well as HuR and tubulin antibodies to confirm efficient fractionation. We achieve robust enrichment of ZC3H14 in each compartment. As expected, HuR and tubulin are present primarily in the nuclear and cytoplasmic fractions, respectively. D, total RNA isolated from the ZC3H14 RNA-IP in each compartment was subjected to qRT-PCR analysis with GAPDH, ATP5G1, and ZC3H14 primers. mRNA levels in the ZC3H14-bound fraction of each compartment were normalized to input levels and then compared by fold-enrichment over pre-immune control. Significant enrichment of GAPDH, ATP5G1, and ZC3H14 mRNAs was observed in the nucleus; however, ZC3H14 was the only transcript significantly enriched in the cytoplasmic samples. Data points represent the mean ± S.E. for n = 3 independent experiments. *, ***, and **** represent p ≤ 0.05, p ≤ 0.001, and p ≤ 0.0001, respectively.
To determine whether the steady-state level of ATP5G1 mRNA is decreased in a specific compartment upon ZC3H14 knockdown, we collected RNA from the nuclear and cytoplasmic samples in Fig. 5A and performed qRT-PCR analyses to detect the GAPDH, ZC3H14, and ATP5G1 transcripts. As shown in Fig. 5B, steady-state ATP5G1 mRNA levels decrease in both the nuclear and cytoplasmic compartments, suggesting that the effect of ZC3H14 knockdown of ATP5G1 mRNA levels is, at least in large part, a nuclear event leading to depletion of nuclear transcripts. As expected, ZC3H14 mRNA levels decrease significantly upon knockdown with no change in the steady-state levels of the control GAPDH transcript.
Given the decrease in ATP5G1 mRNA in the nucleus detected upon ZC3H14 knockdown, we predict that ZC3H14 binds and regulates the ATP5G1 transcript in the nucleus. To determine whether ZC3H14 binds preferentially to ATP5G1 mRNA in the nucleus and/or cytoplasm of MCF-7 cells, we performed ZC3H14 RNA-IP analyses on fractionated nuclear and cytoplasmic lysates. Fig. 5C shows efficient fractionation of these cells, as evidenced by the primarily cytoplasmic and nuclear localization of tubulin and HuR, respectively. We also detect significantly more ZC3H14 in the nuclear samples than in the cytoplasmic samples, as expected (13). Despite the difference in ZC3H14 levels between the two compartments, ZC3H14 is robustly enriched in the bound fraction from each compartment, with no ZC3H14 enrichment in the pre-immune control samples (Fig. 5C).
RNA isolated from input and bound fractions within each compartment was subjected to qRT-PCR analyses to detect GAPDH, ATP5G1, and ZC3H14. As shown in Fig. 5D, ATP5G1 mRNA is significantly enriched upon purification of ZC3H14 in the nucleus but not in the cytoplasm. GAPDH mRNA is also modestly, albeit significantly, enriched in the nuclear fraction only. Interestingly, ZC3H14 mRNA is robustly enriched in both the nuclear and cytoplasmic fractions, suggesting that the mode of ZC3H14 binding to its cognate mRNA is different from other mRNAs examined, potentially via interactions with the conserved internal polyadenosine sequence in the 3′UTR. Together, these data suggest that ZC3H14 interacts with the ATP5G1 transcript specifically in the nucleus and that ZC3H14 may play a role in modulating nuclear processing events involved in the maturation of ATP5G1 mRNA.
ZC3H14 Interacts Preferentially with ATP5G1 Pre-mRNA and Ensures Proper Pre-mRNA Processing
The findings that ZC3H14 interacts with the ATP5G1 transcript exclusively in the nucleus and that ATP5G1 is targeted by NMD suggest that ZC3H14 could interact with the ATP5G1 pre-mRNA to ensure proper pre-mRNA processing. To test this idea, we performed qRT-PCR analyses with RPLP0, RPLP0 pre-mRNA, ATP5G1, ATP5G1 pre-mRNA, and ZC3H14 primers on RNA isolated from the input and bound fractions of ZC3H14 RNA-IPs (Fig. 3E). As in all other experiments, the primers to detect ATP5G1 and RPLP0 span an exon/exon boundary and thus specifically detect mature mRNA (Fig. 6B). Consistent with previous analyses, the ATP5G1 and ZC3H14 mRNAs are robustly enriched upon purification of ZC3H14 (Fig. 6A) with no enrichment of the control RPLP0 mRNA. Intriguingly, ATP5G1 and RPLP0 pre-mRNAs are also enriched upon ZC3H14 purification, in fact at significantly higher levels than their respective mature mRNAs (Fig. 6A). This result suggests that ZC3H14 ensures the proper nuclear maturation of a specific subset of target RNAs that includes ATP5G1.
Previous studies performed in budding yeast provide evidence that Nab2 is involved in the generation of export-competent messenger ribonucleoproteins and retention of improperly processed RNAs (26, 29, 30). Consistent with a similar function for ZC3H14, ATP5G1 pre-mRNA levels increase in the cytoplasm upon ZC3H14 knockdown (Fig. 6B), resulting in a significantly decreased nuclear/cytoplasmic ratio of ATP5G1 pre-mRNA in cells depleted of ZC3H14. As expected, no significant changes in the levels or distribution of the control RPLP0 pre-mRNA were observed (Fig. 6B).
Knockdown of ZC3H14 Causes Mitochondrial Fragmentation Phenocopying Loss of ATP5G1
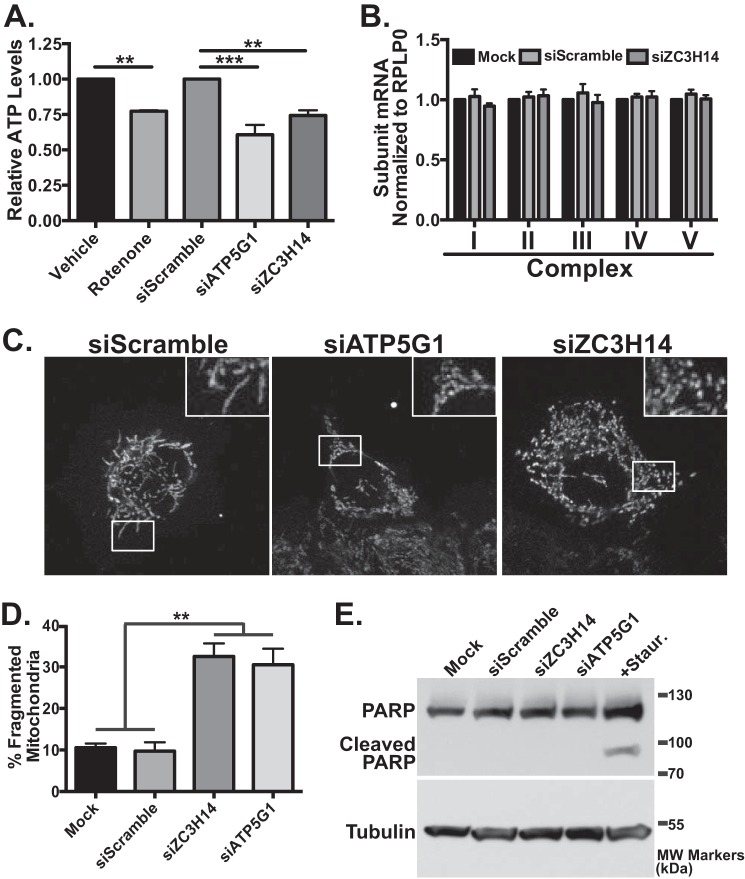
As schematized in Fig. 3A, the mature protein products of the mitochondrial C subunit encoded by the ATP5G genes are identical, preventing us from determining whether knockdown of ZC3H14 results in a specific decrease in the level of ATP5G1 protein. However, previous studies demonstrated that ATP5G1 is rate-limiting for ATP synthase assembly and function and therefore is critical for overall function of the complex and determining cellular ATP levels (64, 65). We hypothesized that if the ZC3H14-dependent reduction of ATP5G1 mRNA results in a decrease in steady-state protein levels, then we should observe a decrease in total cellular ATP levels. To determine whether knockdown of ZC3H14 modulates cellular ATP levels, we performed a cellular ATP extraction followed by a luciferase-based ATP quantification assay (Fig. 7A). As a control, treatment of cells with the electron transport chain inhibitor, rotenone (66), resulted in a significant decrease in cellular ATP levels (Fig. 7A). Cells treated with siRNA targeting ATP5G1 directly or ZC3H14 also displayed significantly decreased cellular ATP levels confirming that ZC3H14 impacts the function of ATP5G1 (Fig. 7A).
FIGURE 7.
Knockdown of ZC3H14 results in fragmented mitochondria. A, to assess cellular ATP levels, cells treated with vehicle control, the electron transport chain inhibitor, rotenone, Scramble siRNA, or siRNA targeting ATP5G1 or ZC3H14 were subjected to boiling water extraction and ATP level quantification using a luciferase-based assay. Cellular ATP levels are normalized to vehicle control or siScramble, which are both set to 1.0 and plotted as relative ATP levels. ZC3H14 knockdown results in decreased cellular ATP levels similar to that observed with rotenone treatment or knockdown of ATP5G1. B, cells treated with a mock transfection, Scramble siRNA, or ZC3H14 siRNA were harvested, and total RNA was used for qRT-PCR analyses. Primers specific to one representative nuclear-encoded mitochondrial mRNA from each OXPHOS complex as well as the control transcript, RPLP0, were used to assess any overall impact of ZC3H14 knockdown on steady-state levels of transcripts encoding OXPHOS components I, NDUFA4; II, SDHB; III, UQCRFS1; IV, CoxIV, and V, ATP5B (67). Relative mRNA values for each OXPHOS mRNA from mock transfection are set to 1.0. C, MCF-7 cells transfected with either Scramble (siScramble), ATP5G1 siRNA (siATP5G1), or ZC3H14 (siZC3H14) were fixed, permeabilized, and subjected to immunofluorescence with cytochrome c antibody. Insets are enlarged from the boxed regions of cells to better highlight mitochondrial morphology. Mitochondrial morphology in cells transfected with scrambled siRNA was indistinguishable from cells transfected with no siRNA (data not shown). D, cells from C were scored for the presence of normal or fragmented mitochondria. Data are represented as a mean averaged from three independent experiments (n = 304 for mock transfected cells, 311 for scrambled siRNA, 307 for ZC3H14 siRNA, and 307 for ATP5G1 siRNA). The difference between control cells treated with Scramble siRNA and either ZC3H14 or ATP5G1 siRNA-treated cells was statistically significant. E, MCF-7 cells treated with mock transfection, Scramble, ZC3H14, or ATP5G1 siRNA or the apoptotic inducer staurosporine (Staur.) were subjected to immunoblot analysis with PARP or tubulin antibody. The presence of cleaved PARP product only in the staurosporine-treated samples suggests that the other cell populations are not undergoing apoptosis. Values represent the mean ± S.E. for n = 3 independent experiments. ** represents p ≤ 0.01; ***, p ≤ 0.01. Images are representative of n = 3 independent experiments with at least 100 cells per experiment in each treatment group.
To extend our analysis to other electron transport chain, or oxidative phosphorylation (OXPHOS) machinery mRNAs, we compared control cells treated with Scramble siRNA to cells depleted for ZC3H14 and then performed qRT-PCR with primers specific to a set of transcripts that represent nuclear-encoded mitochondrial proteins from each of the five OXPHOS complexes (I, NDUFA4; II, SDHB; III, UQCRFS1; IV, CoxIV; and V, ATP5B) (67). As shown in Fig. 7B, no change in steady-state levels of the OXPHOS subunit mRNAs examined was detected, supporting the idea that ZC3H14 does not play a general role in regulating OXPHOS or mitochondrial mRNAs.
A previous study demonstrated that decreasing the steady-state level of ATP5G1 mRNA results in altered mitochondrial morphology (68), consistent with the idea that proper mitochondrial health and function are critical for normal mitochondrial morphology (69, 70). These data, together with our observation that knockdown of ZC3H14 results in a significant decrease in steady-state ATP5G1 levels and cellular ATP levels, suggest that the loss of ZC3H14 could affect mitochondrial morphology. To assess mitochondrial morphology, we transfected cells with siScramble, siATP5G1, or siZC3H14. After 72 h, we fixed cells and stained for either cytochrome c (a mitochondrial intermembrane space protein (71)) or HSP60 (a mitochondrial matrix protein (72)). Control cells either treated with no siRNA (data not shown) or treated with scrambled siRNA (Fig. 7C, left panel) exhibited normal mitochondria with a tubular threadlike morphology. In agreement with previous work (52), we observed a fragmented mitochondrial phenotype in cells transfected with ATP5G1 siRNA (Fig. 7C, middle panel). Consistent with the observation that ZC3H14 decreases the steady-state level of ATP5G1, we also observed a similar degree of mitochondrial fragmentation in cells treated with siZC3H14 (Fig. 7C, right panel). For quantification, cells were scored on the basis of normal mitochondria (predominantly tubular) or fragmented mitochondria (predominantly spherical). As quantified in Fig. 7D, samples transfected with either ZC3H14 or ATP5G1 siRNA had three times as many cells with fragmented mitochondria as control cells, representing 30% of the cell population. We estimate an ∼60% transfection efficiency in the MCF7 cells, so this 30% represents roughly half of transfected cells that show altered mitochondrial morphology. Therefore, we conclude that loss of ZC3H14 causes mitochondrial fragmentation and phenocopies the loss of ATP5G1.
Mitochondrial fragmentation is commonly observed in cells undergoing apoptosis (73). To determine whether knockdown of ATP5G1 or ZC3H14 induces apoptosis causing the observed mitochondrial fragmentation phenotype, we analyzed levels of cleaved PARP. PARP is cleaved during programmed cell death (74) and is therefore a marker for apoptosis. As shown in Fig. 7E, treatment of cells with staurosporine, a protein kinase C inhibitor and potent inducer of apoptosis (75), resulted in PARP cleavage. However, treatment of cells with mock transfection, siScramble, siZC3H14, or siATP5G1 caused no detectable PARP cleavage product, showing that cells treated with these siRNAs are not undergoing apoptosis. Furthermore, no evidence of cytochrome c release (another marker of cells undergoing apoptosis (76)) was observed in the mitochondrial analysis of siRNA-treated cells (Fig. 7C), providing additional support that knockdown of ZC3H14 or ATP5G1 does not trigger apoptosis. Together, these results suggest that loss/reduction of ZC3H14 results in decreased ATP5G1 mRNA levels, ultimately leading to decreased cellular ATP levels and mitochondrial fragmentation.
Discussion
In this study, we performed a genome-wide analysis to assess the impact of ZC3H14 depletion on the steady-state levels of mRNA transcripts. Compared with cells treated with PABPN1 siRNA, which displayed a modulation (≥1.5-fold increase or decrease) in the steady-state level of ∼17% of all expressed transcripts, knockdown of ZC3H14 leads to a significant change in the steady-state level of only ∼1% of all expressed transcripts. This result was surprising given that, like PABPN1, ZC3H14 is a polyadenosine RNA-binding protein (15, 20). There are several possible interpretations of the finding that the steady-state level of a rather smaller number of polyadenylated mRNAs is impacted upon depletion of ZC3H14. First, ZC3H14 could impact gene expression in a manner that does not alter steady-state polyadenylated mRNA levels. For instance, ZC3H14 could modulate the localization and/or translation of target transcripts. The microarray employed also would not detect changes in mRNA export from the nucleus or even splicing within the nucleus. Although the microarray platform employed can detect some common splice variants of certain genes, we are limited in the splicing events that we can observe upon depletion of ZC3H14. Second, depletion of ZC3H14 could impact RNA transcripts not present on the microarray platform such as long noncoding RNAs or miRNAs. Finally, despite biochemical evidence for high affinity binding specifically to polyadenosine RNA (15), ZC3H14 could achieve transcript specificity either through different modes of binding or via interactions with sequence-specific RNA-binding proteins. To address these questions, specific targets of ZC3H14 must be identified and examined.
There are a number of candidate targets identified from the microarray analysis that show a change in the steady-state level upon knockdown of ZC3H14, suggesting that ZC3H14 may regulate the transcription, processing, and/or stability of this subset of mRNAs. To understand how a polyadenosine RNA-binding protein such as ZC3H14 could impact specific target mRNAs, we selected the ATP5G1 transcript for further analysis. The steady-state level of ATP5G1 mRNA is robustly decreased upon knockdown of ZC3H14 but not upon knockdown of PABPN1, suggesting that this transcript is a specific target of ZC3H14.
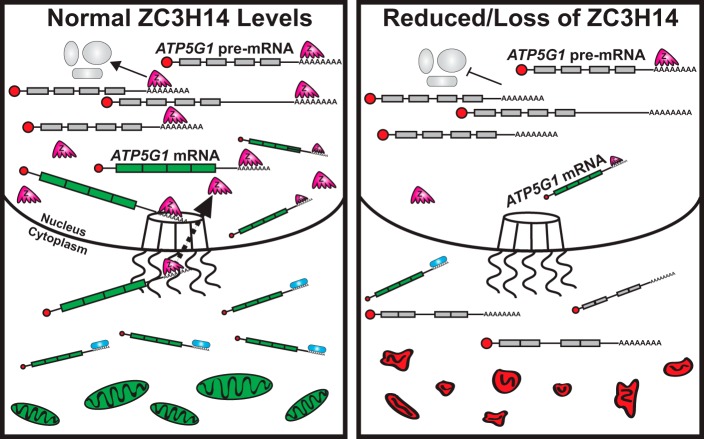
The data presented in this study support a model (Fig. 8, left panel) where ZC3H14 interacts with pre-mRNAs in the nucleus, possibly via interactions with the poly(A) tail, and ensures proper post-transcriptional processing of specific pre-mRNAs (such as ATP5G1). The coupling of these processing events leads to the generation of export-competent mRNPs and prevents aberrantly/incompletely processed mRNAs from exiting the nucleus. In cells with reduced levels of ZC3H14 (Fig. 8, right panel), ZC3H14 is not present to ensure proper processing of target mRNAs, resulting in decreased production of mature target mRNAs (such as ATP5G1) and an increase in improperly/incompletely processed mRNAs in the cytoplasm. As depicted in the model, the decreased level of properly processed, export-competent mRNAs such as ATP5G1 results in altered cellular respiration and defects in mitochondrial morphology.
FIGURE 8.
Model. Left, in cells with normal levels of ZC3H14 (Z; pink, five-fingered shape), ZC3H14 interacts with poly(A) tails throughout nuclear processing events to ensure the coordination of proper pre-mRNA (represented with gray exons and including introns) processing events and to couple these events to export, resulting in the selective export of export-competent mRNPs (represented with green, spliced exons). ZC3H14 is likely removed during the process of export (black dotted arrow). The proper production and export of ATP5G1 mRNA maintains a healthy pool of mitochondria (green ovals at bottom of image). Right, in cells with reduced ZC3H14 levels, post-transcriptional processing events are not properly coordinated, resulting in a decrease in the production of mature mRNA and an increase in improperly and/or incompletely processed pre-mRNAs in the cytoplasm and a disruption in normal mitochondrial morphology (red shapes at bottom of image).
Extensive biochemical studies demonstrate that ZC3H14/Nab2 binds polyadenosine RNA with high affinity (15) and plays an evolutionarily conserved role in poly(A) tail length regulation (12, 24, 31), supporting a model where ZC3H14 likely binds to the poly(A) tails added to virtually all mRNA transcripts. However, the results from our microarray analysis suggest that ZC3H14 could regulate specific mRNA targets in MCF-7 cells. Motif analyses performed on 3′UTR sequences of the 171 transcripts affected by ZC3H14 knockdown reveal a significant representation of A-rich stretches. However, additional studies will be required to determine whether these sequences are enriched within these transcripts relative to the transcriptome as well as whether ZC3H14 could interact with these sequences. Consistent with previous work on S. cerevisiae Nab2, we detected ZC3H14 binding to its cognate mRNA in both the nuclear and cytoplasmic compartments of MCF-7 cells (Fig. 5D). ZC3H14 binding to other transcripts analyzed in the fractionation/RNA-IP experiment was detected only in the nucleus, suggesting that ZC3H14 may interact with its cognate transcript in a distinct manner compared with the other transcripts analyzed, perhaps via the A15 sequence present in the ZC3H14 3′UTR.
There are a number of different models for how ZC3H14 could interact with and impact a specific subset of mRNA transcripts. ZC3H14 could have protein binding partners that provide specificity for target transcripts. For instance, the cytoplasmic Pab, PABPC1, interacts with the human antigen R (HuR) protein, and this interaction is required for the specific stabilization of the β-casein mRNA (77). Our results suggest that ZC3H14 binds preferentially to pre-mRNA transcripts, so perhaps ZC3H14 has binding partners that interact specifically with pre-mRNAs and confer transcript specificity. Alternatively, ZC3H14 could achieve specificity by binding to polyadenosine sequences other than the poly(A) tail. A recent transcriptome-wide analysis revealed that internal, templated polyadenosine stretches are present in a significant number of mRNA transcripts, predominantly in intronic and 3′UTR sequences (20), and therefore could confer specificity to Pabs for target transcripts that contain such additional A-rich sequences. In fact, a recent co-crystal structure of Chaetomium thermophilum Nab2 bound to polyadenosine RNA reveals an RNA-binding module composed of three CCCH zinc fingers that interact with varying numbers of adenosine residues (78). Given that at most two adjacent adenosines can be visualized in this complex of Nab2 zinc fingers bound to oligo(A) (78), Nab2/ZC3H14 could bind to sequences with precise spacing between adenosines and recognize a motif other than homopolymeric adenosine that has not yet been delineated. Consistent with this idea, S. cerevisiae Nab2 can be detected associated with chromatin throughout the genome (30). Further studies will be required to distinguish between these potential modes for ZC3H14 to mediate transcript-specific effects.
Analysis of whole cell lysates across an actinomycin D time course reveals a modest decrease in ATP5G1 mRNA stability upon knockdown of ZC3H14 (Fig. 4C). Typically such effects reflect a change in the cytoplasmic pool of mRNA, suggesting a role for RNA-binding proteins in the cytoplasm. In fact, we do detect a small pool of ZC3H14 isoforms 1–3 in the cytoplasm (Fig. 5A). However, introduction of isoform 1 but not the cytoplasmic isoform (Iso4) rescues ATP5G1 mRNA levels (Fig. 2), suggesting a nuclear role for ZC3H14 in modulating this target mRNA. These observations led us to investigate compartment-specific functions of ZC3H14 (Figs. 5 and 6) as well as interplay between ZC3H14 and the NMD machinery (Fig. 4H).
The results from the fractionation studies not only demonstrate that ZC3H14 binds to the ATP5G1 transcript in the nucleus but also provide evidence for a role in ensuring proper pre-mRNA processing of target transcripts prior to exiting the nucleus. These findings are consistent with a recent study in Schizosaccharomyces pombe that demonstrates a role for the fission yeast ortholog of ZC3H14, Nab2, in acting at the level of the unspliced pre-mRNA and preventing degradation by the nuclear exosome to ensure proper maturation of target transcripts (79). We hypothesize that the inability to ensure the proper nuclear processing of the ATP5G1 pre-mRNA results in a significant decrease in the levels of mature ATP5G1 mRNA in the cytoplasm due to rapid degradation of improperly processed ATP5G1 mRNA via NMD. Consistent with this model, we observe a significant rescue of ATP5G1 mRNA levels upon double knockdown of ZC3H14 and UPF1, a key NMD factor (Fig. 4H). Together, these results suggest that ZC3H14 ensures the proper processing and regulated export of ATP5G1 mRNA from the nucleus. In cells with reduced levels of ZC3H14, improperly processed transcripts such as ATP5G1 are rapidly engaged and degraded by NMD, leading to a significant decrease in mature mRNA and subsequent protein levels. The observed decrease in ATP5G1 steady-state levels correlates with decreased cellular ATP levels and a striking mitochondrial fragmentation phenotype, phenocopying loss of ATP5G1.
The protein product of ATP5G1 is critical for maintaining cellular energy levels, as evidenced by the fact that ATP synthase function is determined by ATP5G1 levels in certain tissues (65). ATP5G1 is the most evolutionarily divergent of the three C subunit genes (53) and is regulated differently from ATP5G2 and ATP5G3 at the transcriptional level (53) as well as in response to various stimuli (80). Aside from this study, the only other work examining the post-transcriptional processing of the ATP5G1 transcript is from studies performed in rat neuronal cells, which demonstrate that the ATP5G1 transcript is localized to and locally translated in axonal tips (68) and is a functional target of miR-338 (81). Loss of ATP5G1 expression specifically in axons resulted in decreased ATP levels, increased reactive oxygen species, and an attenuation of axonal growth rates (68), suggesting that ATP5G1 plays a key role in maintaining proper energy metabolism in neurons. The data presented in this study provide evidence for an additional mechanism that cells employ to ensure tight regulation of ATP5G1 and, in turn, proper ATP production. Furthermore, this study reveals another example of how ATP5G1 is specifically and differentially regulated as compared with the other ATP5G mRNAs and their respective protein products.
The ATP5G1 transcript not only serves as a good target for delineating the role of ZC3H14 in post-transcriptional processing but also provides possible insight into the molecular mechanisms that could contribute to intellectual disability in patients with mutations in ZC3H14 (12). The observed general effect on ATP5G1 detected upon ZC3H14 knockdown in multiple diverse cell lines (including a human neuronal cell line; Fig. 1G) supports the model that ZC3H14 plays a conserved and important role in neurons. Recent work on D. melanogaster Nab2 (dNab2) provides evidence for the critical role of this protein in neurons. Re-expression of dNab2 specifically in the neurons of dNab2 null flies rescues developmental and locomotor defects observed in these mutant flies (12, 31). Importantly, in a trans-species rescue experiment, the nuclear isoform of ZC3H14, isoform 1, but not the cytoplasmic isoform 4, can also rescue dNab2 mutant phenotypes, providing evidence for the critical function of ZC3H14 in neurons (12, 31). The fractionation studies presented here as well as rescue of fly phenotypes by nuclear ZC3H14 are consistent with the model where ZC3H14 is critical for proper maturation of specific target mRNAs within the nucleus.
Properly functioning mitochondria are clearly important in all cell types, but particularly in neurons, which are responsible for the consumption of 25 and 20% of our daily glucose and oxygen, respectively (82, 83). Neurons rely almost exclusively on ATP production via oxidative phosphorylation (84) and are unable to switch to glycolysis when oxidative phosphorylation becomes limited (85). In this study, we found an ∼30% drop in cellular ATP levels with knockdown of either ZC3H14 or ATP5G1. In particular, knockdown of either protein caused a loss of ATP similar to treatment with rotenone, an electron transport chain inhibitor (66). From these data, we conclude that energy production is compromised in cells where ZC3H14 is lost, suggesting a possible respiration defect. In addition, loss of either ZC3H14 or ATP5G1 severely altered mitochondrial morphology, resulting in small, fragmented, round mitochondria. This mitochondrial fragmentation phenotype is frequently observed with loss of electron transport chain function (86–88). Although mitochondrial fragmentation results from an imbalance of mitochondrial fission and fusion and these are coupled to energy metabolism, regulation of these processes remains an active area of research (89, 90). Of note, disruption of normal fission/fusion is observed in many neurological diseases, including dominant optic atrophy, Charcot-Marie-Tooth type 2A, Parkinson's disease, Alzheimer's disease, and Huntington's disease (91). Therefore, we conclude that regulation of ATP5G1 by ZC3H14 is likely to have effects on mitochondria that could be detrimental to neurons.
A major research question is the how defects in or the loss of ubiquitously expressed RNA-binding proteins with general roles in mRNA processing cause tissue-specific disease. For instance, a mutation in the gene encoding the ubiquitously expressed nuclear Pab, PABPN1, causes a specific form of muscular dystrophy that affects primarily ocular and pharyngeal muscles (92). How these specific craniofacial muscles are selectively impacted when all cell types have a requirement for mRNA 3′ end processing and polyadenylation events has yet to be determined. Similarly, patients with loss-of-function mutations in ZC3H14 have a non-syndromic form of intellectual disability (12). Why a loss of ZC3H14 in all tissues would result in a brain-specific phenotype is not at all clear. In fact, many ubiquitously expressed RNA-binding proteins are implicated in neuronal disease (93). Perhaps there are critical binding partners for ZC3H14 found only in neurons. Alternatively, the high energy requirements found in this specialized cell type may make them more vulnerable to fluctuations in energy balance. We propose that ZC3H14, by ensuring the proper pre-mRNA processing of the ATP5G1 transcript, plays a critical role in maintaining proper cellular energy levels and, in turn, mitochondrial health and signaling at neuronal synapses.
Author Contributions
C. P. W. and A. H. C. conceived and coordinated the study. C. P. W. and A. H. C. wrote the paper. C. P. W. performed experiments and analyzed the data. K. J. M. performed and analyzed the experiments shown in Figs. 3D; 4, G and H; and 5C. L. E. N. designed, performed, and analyzed the experiments shown in Fig. 7, C and D. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to the members of the Corbett and Kahn laboratories for helpful discussions and contributions to this work. The HEK293-T17 and HeLa cells were a generous gift from the Kahn laboratory at Emory University. The MB-231 and D556 cells were a generous gift from the Vertino and Read laboratories, respectively, at the Winship Cancer Institute. The Emory Neuroscience Microscopy Core facilities was a recipient of National Institutes of Health Grant P30NS055077.
This work was supported by the National Institutes of Health Grants R01GM058728 (to A. H. C.), R01GM090158 (to R. A. Kahn in support of L. N.), F31CA168321 (to C. P.W.), and T32GM008367 (to C. P.W. and L. N.) and by American Heart Association Grant 14PRE18840040 (to L. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1 and S2.
GEO accession number GSE62067.
- Pab
- poly(A)-binding protein
- NMD
- nonsense-mediated decay
- cNLS
- classical nuclear localization signal
- qRT
- quantitative RT
- FWD
- forward
- REV
- reverse
- IP
- immunoprecipitation
- PARP
- poly(ADP-ribose) polymerase
- ActD
- actinomycin-D
- OXPHOS
- oxidative phosphorylation.
References
- 1. Phillips T. (2008) Regulation of transcription and gene expression in eukaryotes. Nat. Education 1, 199 [Google Scholar]
- 2. Glisovic T., Bachorik J. L., Yong J., and Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moore M. J. (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518 [DOI] [PubMed] [Google Scholar]
- 4. Fasken M. B., and Corbett A. H. (2005) Process or perish: quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 12, 482–488 [DOI] [PubMed] [Google Scholar]
- 5. Blackinton J. G., and Keene J. D. (2014) Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 34, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan Y. A., Hieter P., and Stirling P. C. (2014) Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 30, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukong K. E., Chang K. W., Khandjian E. W., and Richard S. (2008) RNA-binding proteins in human genetic disease. Trends Genet. 24, 416–425 [DOI] [PubMed] [Google Scholar]
- 8. Liu-Yesucevitz L., Bassell G. J., Gitler A. D., Hart A. C., Klann E., Richter J. D., Warren S. T., and Wolozin B. (2011) Local RNA translation at the synapse and in disease. J. Neurosci. 31, 16086–16093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romano M., and Buratti E. (2013) Targeting RNA-binding proteins involved in neurodegeneration. J. Biomol. Screen. 18, 967–983 [DOI] [PubMed] [Google Scholar]
- 10. Wilson A. C., Dugger B. N., Dickson D. W., and Wang D. S. (2011) TDP-43 in aging and Alzheimer's disease–a review. Int. J. Clin. Exp. Pathol. 4, 147–155 [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly S., Pak C., Garshasbi M., Kuss A., Corbett A. H., and Moberg K. (2012) New kid on the ID block: neural functions of the Nab2/ZC3H14 class of Cys(3)His tandem zinc-finger polyadenosine RNA-binding proteins. RNA Biol. 9, 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pak C., Garshasbi M., Kahrizi K., Gross C., Apponi L. H., Noto J. J., Kelly S. M., Leung S. W., Tzschach A., Behjati F., Abedini S. S., Mohseni M., Jensen L. R., Hu H., Huang B., et al. (2011) Mutation of the conserved polyadenosine RNA-binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc. Natl. Acad. Sci. U.S.A. 108, 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leung S. W., Apponi L. H., Cornejo O. E., Kitchen C. M., Valentini S. R., Pavlath G. K., Dunham C. M., and Corbett A. H. (2009) Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA-binding protein. Gene 439, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guthrie C. R., Greenup L., Leverenz J. B., and Kraemer B. C. (2011) MSUT2 is a determinant of susceptibility to tau neurotoxicity. Hum. Mol. Genet. 20, 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelly S. M., Pabit S. A., Kitchen C. M., Guo P., Marfatia K. A., Murphy T. J., Corbett A. H., and Berland K. M. (2007) Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 12306–12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kühn U., Nemeth A., Meyer S., and Wahle E. (2003) The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 278, 16916–16925 [DOI] [PubMed] [Google Scholar]
- 17. Baer B. W., and Kornberg R. D. (1983) The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 96, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kühn U., and Pieler T. (1996) Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 256, 20–30 [DOI] [PubMed] [Google Scholar]
- 19. Sachs A. B., Davis R. W., and Kornberg R. D. (1987) A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7, 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wigington C. P., Williams K. R., Meers M. P., Bassell G. J., and Corbett A. H. (2014) Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip. Rev. RNA 5, 601–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fasken M. B., Stewart M., and Corbett A. H. (2008) Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J. Biol. Chem. 283, 27130–27143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson J. T., Wilson S. M., Datar K. V., and Swanson M. S. (1993) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13, 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilson S. M., Datar K. V., Paddy M. R., Swedlow J. R., and Swanson M. S. (1994) Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soucek S., Corbett A. H., and Fasken M. B. (2012) The long and the short of it: the role of the zinc finger polyadenosine RNA-binding protein, Nab2, in control of poly(A) tail length. Biochim. Biophys. Acta 1819, 546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Green D. M., Marfatia K. A., Crafton E. B., Zhang X., Cheng X., and Corbett A. H. (2002) Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277, 7752–7760 [DOI] [PubMed] [Google Scholar]
- 26. Hector R. E., Nykamp K. R., Dheur S., Anderson J. T., Non P. J., Urbinati C. R., Wilson S. M., Minvielle-Sebastia L., and Swanson M. S. (2002) Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21, 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viphakone N., Voisinet-Hakil F., and Minvielle-Sebastia L. (2008) Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 36, 2418–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Apponi L. H., Kelly S. M., Harreman M. T., Lehner A. N., Corbett A. H., and Valentini S. R. (2007) An interaction between two RNA-binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability. Mol. Cell. Biol. 27, 6569–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soucek S., Z. Y., Bellur Deepti L., Bergkessel Megan, Guthrie Christine, Staley Jonathan P., and Corbett Anita H. (2016) A role for the polyadenosine RNA-binding protein, Nab2, in splicing and RNA surveillance. Mol. Biol. Cell 10.1128/MCB.00402-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tuck A. C., and Tollervey D. (2013) A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154, 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelly S. M., Leung S. W., Pak C., Banerjee A., Moberg K. H., and Corbett A. H. (2014) A conserved role for the zinc finger polyadenosine RNA-binding protein, ZC3H14, in control of poly(A) tail length. RNA 20, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dyer M. R., and Walker J. E. (1993) Sequences of members of the human gene family for the c subunit of mitochondrial ATP synthase. Biochem. J. 293, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jonckheere A. I., Smeitink J. A., and Rodenburg R. J. (2012) Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit. Metab. Dis. 35, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soule H. D., Vazguez J., Long A., Albert S., and Brennan M. (1973) A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 51, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 35. Towbin H., Staehelin T., and Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Apponi L. H., Leung S. W., Williams K. R., Valentini S. R., Corbett A. H., and Pavlath G. K. (2010) Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 19, 1058–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 38. Yang N. C., Ho W. M., Chen Y. H., and Hu M. L. (2002) A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal. Biochem. 306, 323–327 [DOI] [PubMed] [Google Scholar]
- 39. Kühn U., and Wahle E. (2004) Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678, 67–84 [DOI] [PubMed] [Google Scholar]
- 40. Mangus D. A., Evans M. C., and Jacobson A. (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kerwitz Y., Kühn U., Lilie H., Knoth A., Scheuermann T., Friedrich H., Schwarz E., and Wahle E. (2003) Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 22, 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kühn U., Gündel M., Knoth A., Kerwitz Y., Rüdel S., and Wahle E. (2009) Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66, 759–768 [DOI] [PubMed] [Google Scholar]
- 44. Beaulieu Y. B., Kleinman C. L., Landry-Voyer A. M., Majewski J., and Bachand F. (2012) Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 8, e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abouantoun T. J., and MacDonald T. J. (2009) Imatinib blocks migration and invasion of medulloblastoma cells by concurrently inhibiting activation of platelet-derived growth factor receptor and transactivation of epidermal growth factor receptor. Mol. Cancer Ther. 8, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guppy M., Leedman P., Zu X., and Russell V. (2002) Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem. J. 364, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reitzer L. J., Wice B. M., and Kennell D. (1979) Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 254, 2669–2676 [PubMed] [Google Scholar]
- 48. Watt I. N., Montgomery M. G., Runswick M. J., Leslie A. G., and Walker J. E. (2010) Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dyer M. R., Gay N. J., and Walker J. E. (1989) DNA sequences of a bovine gene and of two related pseudogenes for the proteolipid subunit of mitochondrial ATP synthase. Biochem. J. 260, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan W. L., Lerner T. J., Haines J. L., and Gusella J. F. (1994) Sequence analysis and mapping of a novel human mitochondrial ATP synthase subunit 9 cDNA (ATP5G3). Genomics 24, 375–377 [DOI] [PubMed] [Google Scholar]
- 51. Sangawa H., Himeda T., Shibata H., and Higuti T. (1997) Gene expression of subunit c(P1), subunit c(P2), and oligomycin sensitivity-conferring protein may play a key role in biogenesis of H+-ATP synthase in various rat tissues. J. Biol. Chem. 272, 6034–6037 [DOI] [PubMed] [Google Scholar]
- 52. Vives-Bauza C., Magrané J., Andreu A. L., and Manfredi G. (2010) Novel role of ATPase subunit C targeting peptides beyond mitochondrial protein import. Mol. Biol. Cell 21, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Grassi A., Lanave C., and Saccone C. (2006) Evolution of ATP synthase subunit c and cytochrome c gene families in selected Metazoan classes. Gene 371, 224–233 [DOI] [PubMed] [Google Scholar]
- 54. Gay N. J., and Walker J. E. (1985) Two genes encoding the bovine mitochondrial ATP synthase proteolipid specify precursors with different import sequences and are expressed in a tissue-specific manner. EMBO J. 4, 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roth K. M., Wolf M. K., Rossi M., and Butler J. S. (2005) The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol. 25, 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pullmann R. Jr, Kim H. H., Abdelmohsen K., Lal A., Martindale J. L., Yang X., and Gorospe M. (2007) Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 27, 6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hurwitz J., Furth J. J., Malamy M., and Alexander M. (1962) The role of deoxyribonucleic acid in ribonucleic acid synthesis. III. The inhibition of the enzymatic synthesis of ribonucleic acid and deoxyribonucleic acid by actinomycin D and proflavin. Proc. Natl. Acad. Sci. U.S.A. 48, 1222–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 59. Wu X., and Brewer G. (2012) The regulation of mRNA stability in mammalian cells: 2.0. Gene 500, 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karam R., Wengrod J., Gardner L. B., and Wilkinson M. F. (2013) Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim. Biophys. Acta 1829, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Popp M. W., and Maquat L. E. (2013) Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47, 139–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kurosaki T., Li W., Hoque M., Popp M. W., Ermolenko D. N., Tian B., and Maquat L. E. (2014) A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 28, 1900–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fan X. C., and Steitz J. A. (1998) HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U.S.A. 95, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Houstĕk J., Andersson U., Tvrdík P., Nedergaard J., and Cannon B. (1995) The expression of subunit c correlates with and thus may limit the biosynthesis of the mitochondrial F0F1-ATPase in brown adipose tissue. J. Biol. Chem. 270, 7689–7694 [DOI] [PubMed] [Google Scholar]
- 65. Kramarova T. V., Shabalina I. G., Andersson U., Westerberg R., Carlberg I., Houstek J., Nedergaard J., and Cannon B. (2008) Mitochondrial ATP synthase levels in brown adipose tissue are governed by the c-Fo subunit P1 isoform. FASEB J. 22, 55–63 [DOI] [PubMed] [Google Scholar]
- 66. Chance B., Williams G. R., and Hollunger G. (1963) Inhibition of electron and energy transfer in mitochondria. I. Effects of Amytal, thiopental, rotenone, progesterone, and methylene glycol. J. Biol. Chem. 238, 418–431 [PubMed] [Google Scholar]
- 67. Duborjal H., Beugnot R., Mousson de Camaret B., and Issartel J. P. (2002) Large functional range of steady-state levels of nuclear and mitochondrial transcripts coding for the subunits of the human mitochondrial OXPHOS system. Genome Res. 12, 1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Natera-Naranjo O., Kar A. N., Aschrafi A., Gervasi N. M., Macgibeny M. A., Gioio A. E., and Kaplan B. B. (2012) Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol. Cell. Neurosci. 49, 263–270 [DOI] [PubMed] [Google Scholar]
- 69. Karbowski M., and Youle R. J. (2003) Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 10, 870–880 [DOI] [PubMed] [Google Scholar]
- 70. Yaffe M. P. (1999) The machinery of mitochondrial inheritance and behavior. Science 283, 1493–1497 [DOI] [PubMed] [Google Scholar]
- 71. Kaput J., Brandriss M. C., and Prussak-Wieckowska T. (1989) In vitro import of cytochrome c peroxidase into the intermembrane space: release of the processed form by intact mitochondria. J. Cell Biol. 109, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Singh B., Patel H. V., Ridley R. G., Freeman K. B., and Gupta R. S. (1990) Mitochondrial import of the human chaperonin (HSP60) protein. Biochem. Biophys. Res. Commun. 169, 391–396 [DOI] [PubMed] [Google Scholar]
- 73. Suen D. F., Norris K. L., and Youle R. J. (2008) Mitochondrial dynamics and apoptosis. Genes Dev. 22, 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., and Poirier G. G. (1993) Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 53, 3976–3985 [PubMed] [Google Scholar]
- 75. Belmokhtar C. A., Hillion J., and Ségal-Bendirdjian E. (2001) Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 20, 3354–3362 [DOI] [PubMed] [Google Scholar]
- 76. Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., and Wang X. (1997) Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 77. Nagaoka K., Suzuki T., Kawano T., Imakawa K., and Sakai S. (2006) Stability of casein mRNA is ensured by structural interactions between the 3′-untranslated region and poly(A) tail via the HuR and poly(A)-binding protein complex. Biochim. Biophys. Acta 1759, 132–140 [DOI] [PubMed] [Google Scholar]
- 78. Kuhlmann S. I., Valkov E., and Stewart M. (2014) Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers. Nucleic Acids Res. 42, 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Grenier St-Sauveur V., Soucek S., Corbett A. H., and Bachand F. (2013) Poly(A) tail-mediated gene regulation by opposing roles of Nab2 and Pab2 nuclear poly(A)-binding proteins in pre-mRNA decay. Mol. Cell. Biol. 33, 4718–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Andersson U., Houstek J., and Cannon B. (1997) ATP synthase subunit c expression: physiological regulation of the P1 and P2 genes. Biochem. J. 323, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Aschrafi A., Kar A. N., Natera-Naranjo O., MacGibeny M. A., Gioio A. E., and Kaplan B. B. (2012) MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell. Mol. Life Sci. 69, 4017–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kann O., and Kovács R. (2007) Mitochondria and neuronal activity. Am. J. Physiol. Cell Physiol. 292, C641–C657 [DOI] [PubMed] [Google Scholar]
- 83. Silver I., and Erecińska M. (1998) Oxygen and ion concentrations in normoxic and hypoxic brain cells. Adv. Exp. Med. Biol. 454, 7–16 [DOI] [PubMed] [Google Scholar]
- 84. Ames A., 3rd (2000) CNS energy metabolism as related to function. Brain Res. Brain Res. Rev. 34, 42–68 [DOI] [PubMed] [Google Scholar]
- 85. Knott A. B., Perkins G., Schwarzenbacher R., and Bossy-Wetzel E. (2008) Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 9, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koopman W. J., Visch H. J., Verkaart S., van den Heuvel L. W., Smeitink J. A., and Willems P. H. (2005) Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am. J. Physiol. Cell Physiol. 289, C881–C890 [DOI] [PubMed] [Google Scholar]
- 87. Kwong J. Q., Henning M. S., Starkov A. A., and Manfredi G. (2007) The mitochondrial respiratory chain is a modulator of apoptosis. J. Cell Biol. 179, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sauvanet C., Duvezin-Caubet S., di Rago J. P., and Rojo M. (2010) Energetic requirements and bioenergetic modulation of mitochondrial morphology and dynamics. Semin. Cell Dev. Biol. 21, 558–565 [DOI] [PubMed] [Google Scholar]
- 89. Chan D. C. (2012) Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 [DOI] [PubMed] [Google Scholar]
- 90. Youle R. J., and van der Bliek A. M. (2012) Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen H., and Chan D. C. (2009) Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Banerjee A., Apponi L. H., Pavlath G. K., and Corbett A. H. (2013) PABPN1: molecular function and muscle disease. FEBS J. 280, 4230–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lenzken S. C., Achsel T., Carrì M. T., and Barabino S. M. (2014) Neuronal RNA-binding proteins in health and disease. Wiley Interdiscip. Rev. RNA 5, 565–576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.