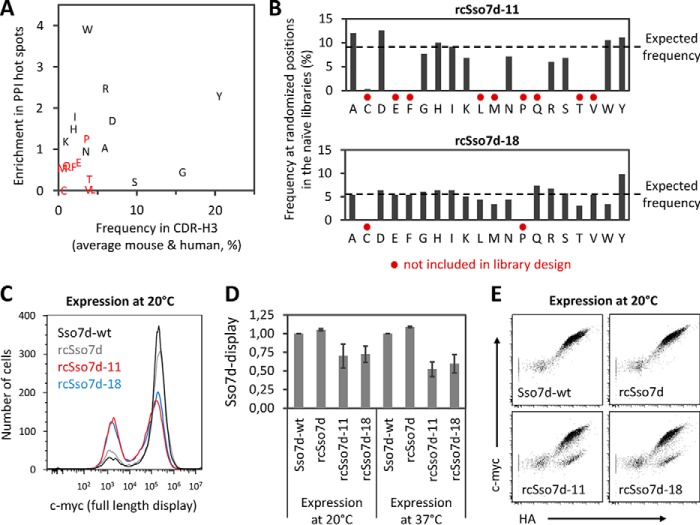
FIGURE 2.
Construction and analysis of rcSso7d libraries. A, enrichments of all amino acids in PPI hot spots (23) are plotted against their frequencies in CDR-H3 loops of murine and human antibodies (24). Only the amino acids in black were included in the rcSso7d-11 library, whereas the rcSso7d-18 library contained all amino acids except for Cys and Pro. B, amino acid distributions at the randomly mutated binding site positions were analyzed by sequencing 39 and 33 randomly picked clones from the naïve libraries rcSso7d-11 and rcSso7d-18, respectively. Amino acids that were not included in the library design are marked with red dots, and the expected frequencies are indicated by a dashed line. C and D, Sso7d-WT, rcSso7d, and the rcSso7d-11 and rcSso7d-18 libraries were expressed on the surface of yeast at 20 or 37 °C followed by detection of full-length Sso7d mutants by using mouse anti-c-MYC. In C, one representative experiment of three is shown (expression at 20 °C). In D, expression levels on c-MYC-positive cells after normalization for Sso7d-WT expression are shown for both 20 and 37 °C expression (error bars represent S.D. of three independent experiments). E, the expression level of full-length Sso7d mutants (c-MYC) was plotted against total surface expression (the HA tag is located between Aga2p and Sso7d) (expression at 20 °C).