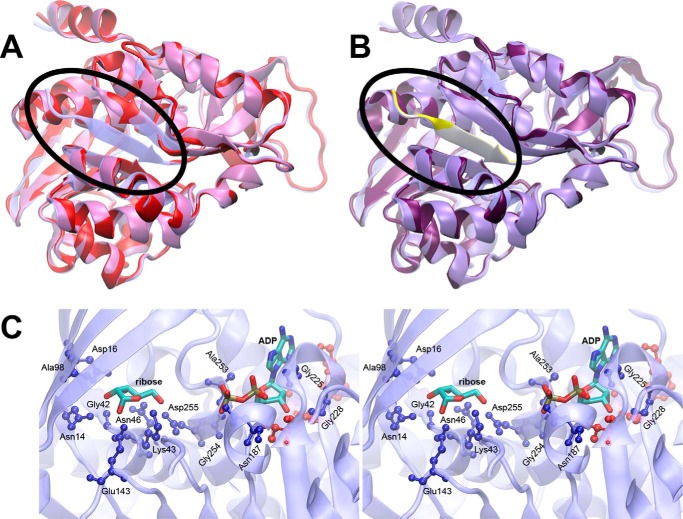
FIGURE 2.
Structure modeling of At1g17160 using E. coli RBSK structure. A, a homology model of At1g17160.1 (red) based on the predicted transit peptide cleavage site is shown superimposed on the template structure of E. coli ribokinase (transparent blue). The N terminus of the E. coli RBSK containing the β-sheet (blue) missing from the At1g17160 model (red) is circled. B, a similar superposition is shown for the homology model with a longer N-terminal sequence (purple), with the added N-terminal sequence highlighted (yellow). C, a stereo view of the E. coli ribokinase active site is shown. Bound ADP and ribose are shown as sticks, colored by atom type. Protein residues within 3.5 Å of ADP or ribose (ball-and-stick) are color-coded by sequence identity to At1g17160 (blue, conserved; red, not conserved).