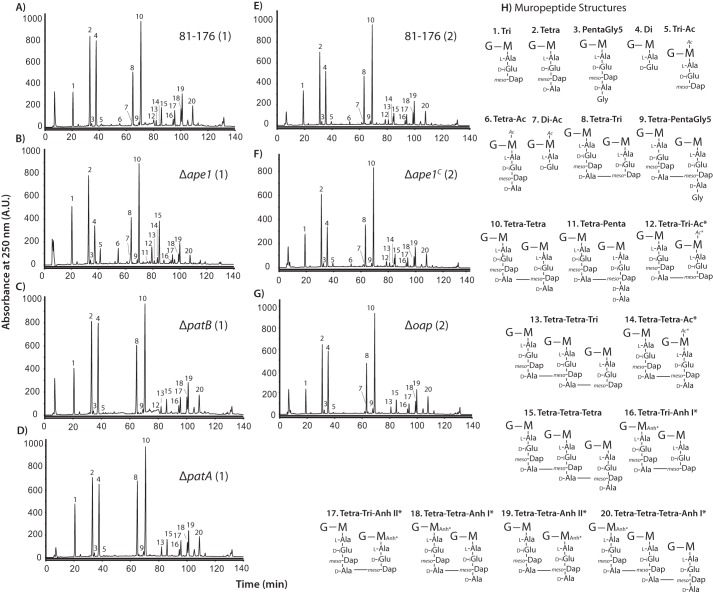
FIGURE 2.
HPLC elution profile of C. jejuni muropeptides and proposed muropeptide structures. Purified PG was digested with cellosyl, and the resulting muropeptides were reduced with sodium borohydride and separated on a Prontosil 120-3-C18 AQ reverse-phase column. HPLC profiles are shown for wild-type strain 81-176 (A and E), Δape1 (B), ΔpatB (C), ΔpatA (D), Δape1C (F), and Δoap (G). Muropeptide profiles were generated in two sets of experiments indicated by (1) for Sample Set 1 and (2) for Sample Set 2. The muropeptide structure represented by each peak was determined previously by mass spectroscopy (18), and proposed muropeptide structures of each peak corresponding to the peak number in the chromatogram are shown in H. The summary of the muropeptide composition is shown in Table 3. G, N-acetylglucosamine; M, reduced N-acetylmuramic acid; l-Ala, l-alanine; d-iGlu, d-isoglutamic acid; meso-Dap, meso-diaminopimelic acid; d-Ala, d-alanine Ac, O-acetyl groups at MurNAc C6 position; Anh, 1,6-anhydro group of MurNAc; *, it is not known on which MurNAc residue the modification occurs.