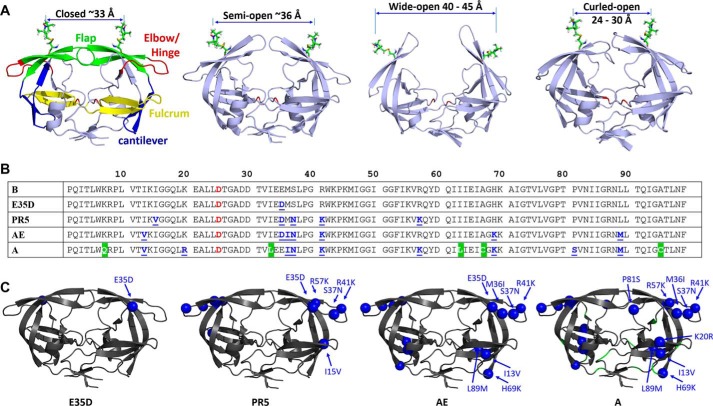
FIGURE 1.
HIV-1 PR conformations and sequences. A, conformations of the four states are shown, “closed (PDB code 2BPX),” “semi-open (PDB code 1TW7),” “wide-open (MD coordinates),” and “curled (MD coordinates),” with 1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl, MTSL, attached to residue 55 using Multiscale Modeling of Macromolecular systems to generate the anticipated distances. Colored segments with residue span in parentheses in the “closed” conformation highlight distinct regions that are referred to throughout the text: flaps (green, residues 43–59), elbow/hinge (red, residues 34–42), fulcrum (yellow, residues 9–24), and cantilever (blue, residues 60–74). B, amino acid sequences of HIV-1 PR variants B, E35D, PR5, CRF01_AE, and subtype A constructs where the catalytic residue (Asp-25) is highlight in red and natural polymorphisms are underlined in blue. All sequences except for A contain the three stabilizing mutations and CYS substitutions (Q7K, L33I, L63I, C67A, and C95A), where alternative sequences for subtype A are shown in a green background. Note for x-ray studies, EPR, and NMR measurements, the substitution D25N was included to render the enzyme inactive. C, locations and identities of natural polymorphisms of each construct are shown as spheres on HIV-1 PR (PDB code 2PK5 (86)).