FIGURE 4.
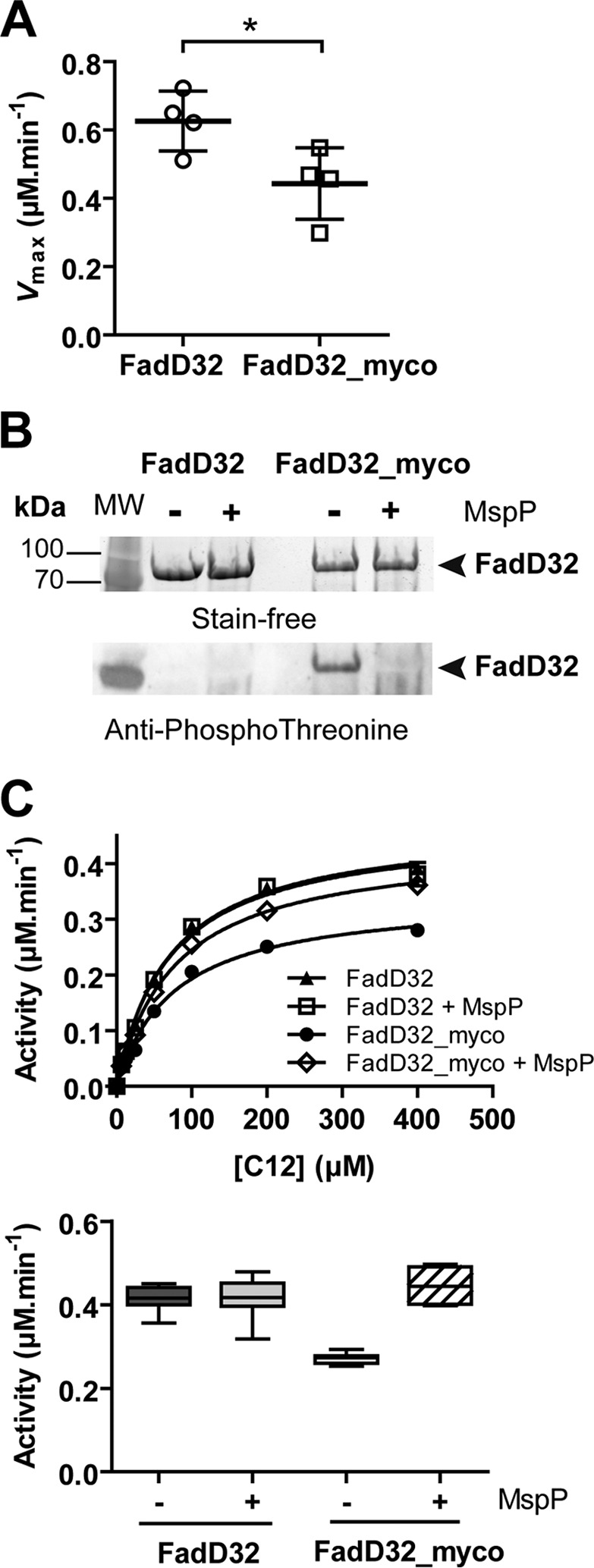
Phosphorylation status and activity of FadD32 produced in mycobacteria. The FadD32 protein was produced either in E. coli (non-phosphorylated, FadD32) or in M. smegmatis (FadD32_myco). A, scatter plots showing maximum velocity Vmax for lauric acid (C12); values are representative of four sets of experiments with independent protein preparations ± S.D. The p value = 0.036 was calculated by Student's t test. B, SDS-PAGE and immunoblot analysis of FadD32 and FadD32_myco, treated or not with purified MspP in vitro. The proteins are detected by Stain-FreeTM SDS-PAGE technology (Bio-Rad) (upper panel), and their phosphorylation status was analyzed by immunoblot using anti-phosphothreonine antibodies (lower panel). C, FAAL activity of FadD32 and FadD32_myco, with or without MspP treatment, was assessed spectrophotometrically in the inorganic pyrophosphate-coupled assay. The Michaelis-Menten non-linear regression curves are displayed using C12 as substrate; values are means ± S.E. of four experimental replicates (upper panel). Lower panel, whisker plots showing activity (reaction velocity at 0.4 μm FadD32, 400 μm C12, and 2 mm ATP) of FadD32_myco with or without MspP treatment; the non-phosphorylated FadD32 proteins treated or not with MspP are shown as a control. Values represent eight experimental replicates issued from two sets of experiments with independent protein preparations; whiskers, minimum to maximum.
