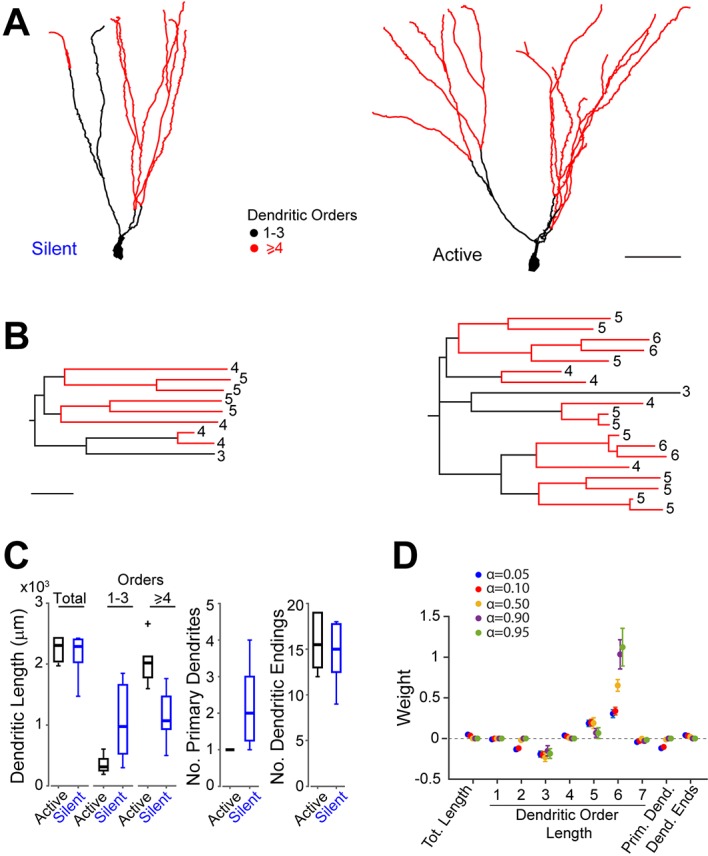
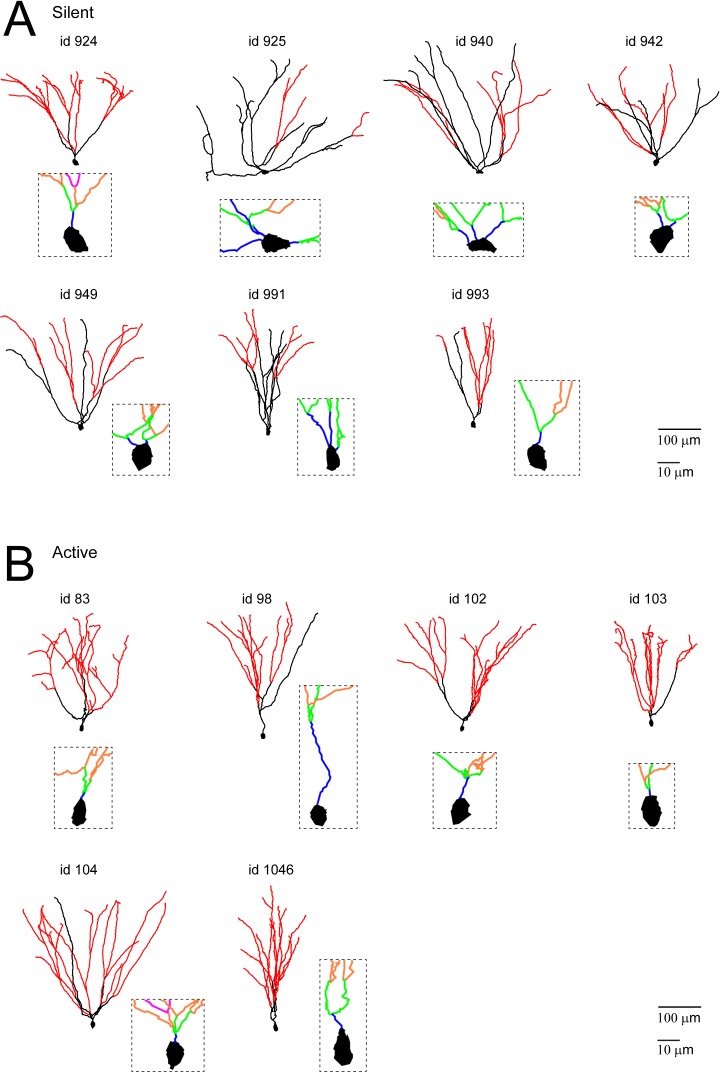
Figure 4. Morphological analysis of identified active and silent granule cells.
(A) Somatodendritic reconstructions of silent (left, cell id 993) and active (right, cell id 102) GCs, recorded in freely moving rats. Dendritic branch orders 1–3 are indicated in black, while high-order branches (≥ 4) are indicated in red. Scale bar=50 µm. (B) Dendrograms for the cells in (A). The branch orders of the terminal dendritic tips are indicated. Color codes same as in (A). Scale bar=50 µm. (C) Box-plots comparing primary dendritic parameters (active GCs, n=6; silent GCs, n=7). Whiskers represent 1.5 IQR. Outliers are shown as crosses. (D) Weights of logistic regression classifier (mean ± SEM over cross-validation folds) used for classifying GCs as active or silent for different levels of sparseness (α=0.05 corresponds to dense weights, α=0.95 corresponds to sparse weights).
DOI: http://dx.doi.org/10.7554/eLife.20252.007