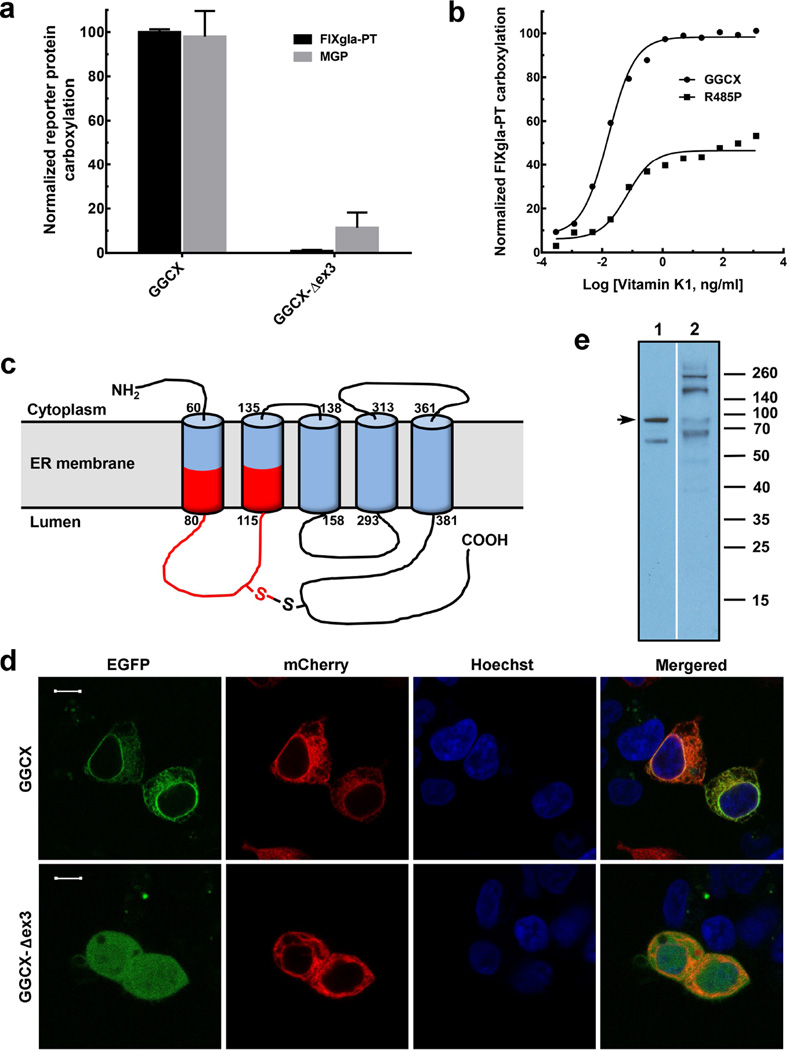
Figure 1. Characterization of GGCX mutations identified in PXE-like and VKCFD patients.
(a) Cell-based activity assay of GGCX and its mutant identified from PXE-like patients. Wild-type GGCX and the GGCX-Δex3 mutant were transiently expressed in GGCX-deficient HEK293 reporter cells. Transfected cells were cultured in complete medium containing 5 µg/ml vitamin K. The concentrations of the carboxylated reporter proteins (FIXgla-PT, black bars and MGP, gray bars) in the cell culture medium were measured by ELISA. Wild-type GGCX activity was normalized to 100%. Data are presented as mean±SD (n=3). (b) Carboxylation activity of GGCX and the Arg485Pro mutant in response to increasing concentrations of vitamin K. Wild-type GGCX (filled circles) and the Arg485Pro mutant (filled squares) were transiently expressed in GGCX-deficient HEK293 reporter cells. The enzymatic activity for FIXgla-PT carboxylation was determined as described above. (c) Schematic representation of the proposed membrane topology of GGCX. The exon 3 encoded region is shown in red. (d) Subcellular localization of GGCX and the GGCX-Δex3 mutant in HEK293 cells. EGFP-tagged GGCX or GGCX-Δex3 was transiently co-expressed with the ER marker mCherry-ER-3 in HEK293 cells. Forty-eight hours post-transfection, cell nucleus was stained by Hoechst 33342 and directly used for fluorescence confocal image collection. GGCX fusions were visualized by the green fluorescence of EGFP, ER marker was visualized by the red fluorescence of mCherry, and cell nuclei were visualized by the blue fluorescence of Hoechst. Scale bar = 10 µm. (e) Immunoblotting analysis of GGCX (lane 1) and the GGCX-Δex3 mutant (lane 2) proteins. Full-length GGCX band is indicated by arrow.