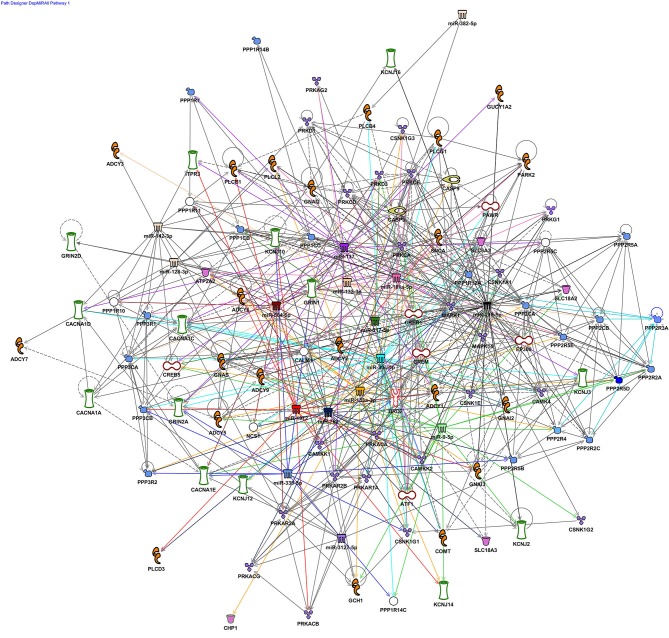
Figure 1.
Dopaminergic miRNA nodes in a rich network of connections in synaptic plasticity pathways. Ingenuity pathway analysis (IPA) was used to explore the molecular connectivity predicted between miRNA implicated in dopaminergic development, function and disease (references for all implicated miRNAs examined in this figure are listed in Table 1). miRNA targets, both experimentally validated and predicted by TargetScan were filtered with respect to dopamine-associated canonical pathways and used to generate a network graph. Core miRNA nodes near the center of the graph are color-coded and have colored edges to denote their connections. The largest network defined by the filter was Dopamine-DARPP32 in cAMP Signaling (85 genes) followed by Dopamine Receptor Signaling (40). These networks were also enriched with synaptic plasticity pathways, Synaptic LTP/LTD (43/29 genes), CREB Signaling in Neurons (43 genes), and the Axon Guidance Signaling (27 genes).