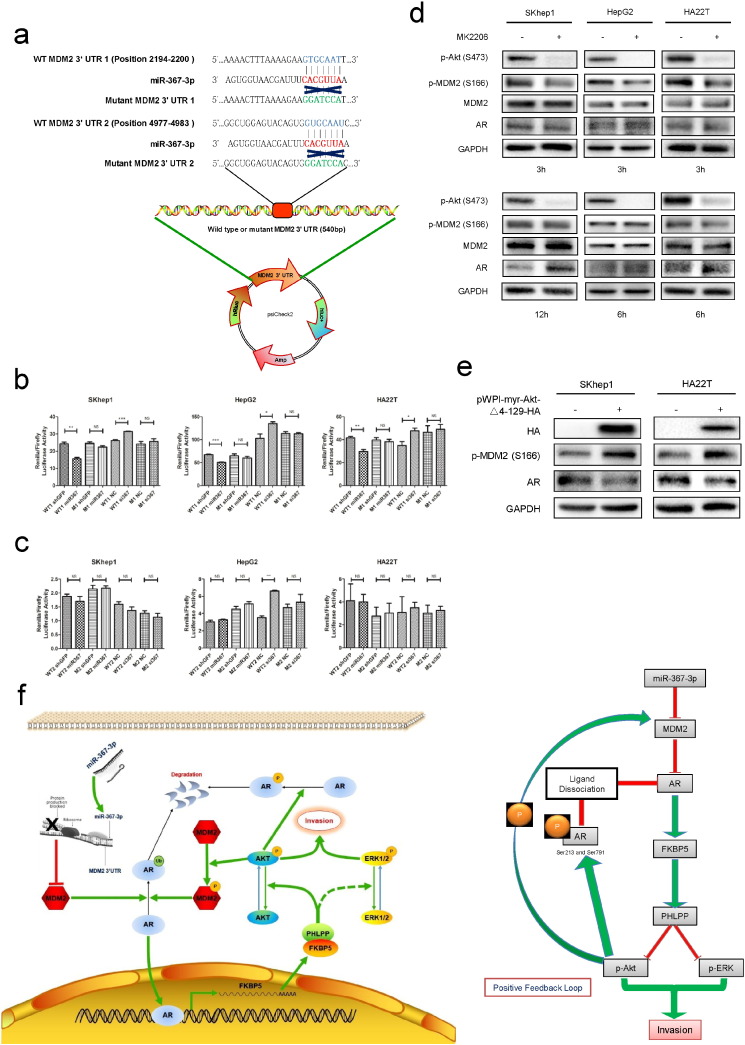
Fig. 5.
The miR-367-3p directly targets the 3′UTR of MDM2 to mediate AR levels and subsequently generates a positive feedback loop.
(a), MDM2 3′UTR containing wild type (WT) or mutant (M) miRNA-response elements were cloned into the psiCheck2 construct downstream of the Renilla luciferase ORF, (b), co-transfection of MDM2 3′UTR constructs containing WT seed regions with pLKO.1-miR-367-3p decreased the luciferase activity and with miR-367-3p inhibitor increased the luciferase activity, whereas the reporter with a M seed region in MDM2 3′UTR did not, in the first predicted miR-367-3p seed regions. (c), co-transfection of MDM2 3′UTR constructs containing WT seed regions with pLKO.1-miR-367-3p or with miR-367-3p inhibitor (50 nM) could not alter the luciferase activity, neither did the reporter with a M seed region in MDM2 3′UTR, in the second predicted miR-367-3p seed regions. (d), western blot assays showed that MK2206 (1 μM in SKhep1 and HA22T cells, 2.5 μM in HepG2 cells) suppressed MDM2 phosphorylation and increased the AR protein expression at various time points. (e), over-expressing pWPI-myr-Akt-Δ4-129-HA in HCC cells showed significant increases of pMDM2 (S166) and decreases of AR. (f), the schematic depiction showing the suppression of pAkt by miR-367-3p could subsequently attenuate the phosphorylation of AR and MDM2, giving rise to additionally increased AR protein levels, which formed a positive feed-back circuitry. p < 0.05 was considered statistically significant. * p < 0.05, ** p < 0.01, and *** p < 0.001.